Single cell Bioconductor's choice: SCE
The rapid advancement of single-cell RNA sequencing (scRNA-seq) has provided unparalleled insights into cellular heterogeneity. However, managing and analyzing the increasing amounts of single-cell data requires robust computational frameworks adapted to this purpose.
Three main options are available:
- Scanpy from the Theis Lab in Python
- SingleCellExperiment (SCE) from Bioconductor in R
- Seurat from Satija Lab in R
In this blog post, I will introduce SingleCellExperiment (SCE), a core Bioconductor framework for handling single-cell data in R. Whether you are exploring scRNA-seq for the first time or transitioning to Bioconductor-based workflows, this guide will walk you through essential concepts and practical applications.
In this post we will use the same dataset of blood cells from the cellxgene database that we used in the Scanpy and Seurat posts.
However, the cellxgene database does not provide the data in a format that can be directly used by the SCE package. Therefore, we will convert the object from Seurat to SCE format (see below).
We start loading the object we used in the Seurat post
1library(Seurat)
2rds.path <- '/media/alfonso/data/velocity_MGI/'
3rds.file <- file.path( rds.path, '582149d8-2a8f-44cf-9605-337b8ca8d518.rds' )
4seurat <- readRDS(rds.file)
check the object
1seurat
1## An object of class Seurat
2## 61759 features across 85233 samples within 1 assay
3## Active assay: RNA (61759 features, 0 variable features)
4## 2 layers present: counts, data
5## 7 dimensional reductions calculated: pca, scvi, tissue_uncorrected_umap, umap, umap_scvi_full_donorassay, umap_tissue_scvi_donorassay, uncorrected_umap
SingleCellExperiment
SingleCellExperiment or SCE is the Bioconductor's R package designed to store and manipulate single cell data. An sce object has the following structure:
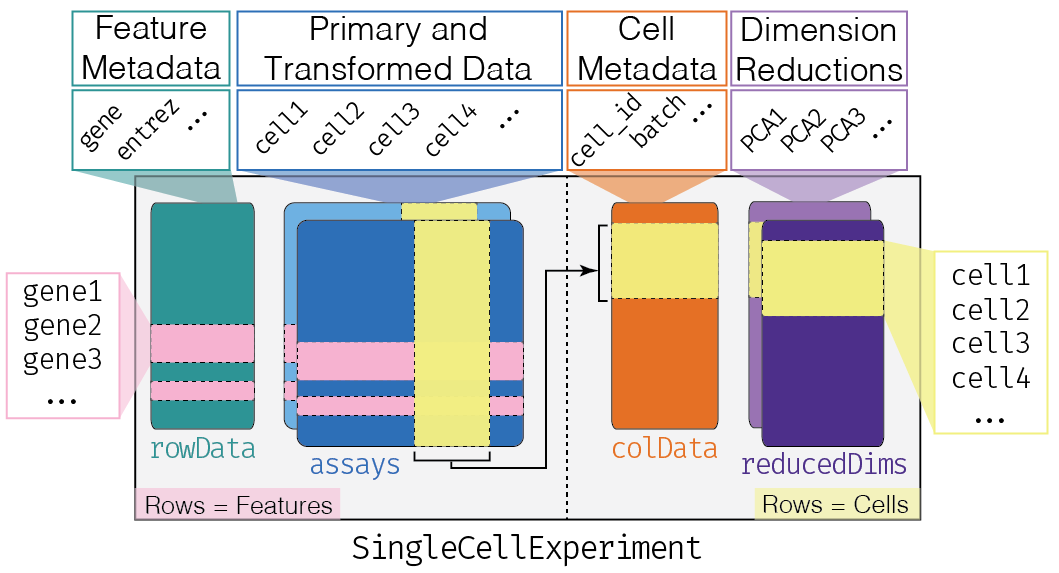
The main advantage of being part of the Bioconductor project is that we can directly use functions from different Bioconductor packages without format conversion. Thus, using SCE provides direct access to 70+ single-cell-related Bioconductor packages.
We convert the Seurat's object to SCE using as.SingleCellExperiment Seurat's function.
1sce <- as.SingleCellExperiment(seurat)
Now we can inspect the data.
1sce
1## class: SingleCellExperiment
2## dim: 61759 85233
3## metadata(0):
4## assays(2): counts logcounts
5## rownames(61759): ENSG00000000003 ENSG00000000005 ... ENSG00000290165
6## ENSG00000290166
7## rowData names(0):
8## colnames(85233): LinNeg_G15 LinNeg_L11 ...
9## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG
10## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG
11## colData names(56): donor_id tissue_in_publication ... nFeature_RNA
12## ident
13## reducedDimNames(7): PCA SCVI ... UMAP_TISSUE_SCVI_DONORASSAY
14## UNCORRECTED_UMAP
15## mainExpName: RNA
16## altExpNames(0):
It is a SingleCellExperiment object with 61759 rows and 85233 columns with two assays (more on this later). The samples (columns) are the cells and the features (rows) are the genes.
Cell metadata
We can explore the cell-related information using the colData function. To use SCE functions we must load the package first.
1library(SingleCellExperiment)
1colData(sce)
1## DataFrame with 85233 rows and 56 columns
2## donor_id tissue_in_publication
3## <factor> <factor>
4## LinNeg_G15 TSP2 Blood
5## LinNeg_L11 TSP2 Blood
6## LinNeg_J16 TSP2 Blood
7## LinNeg_F5 TSP2 Blood
8## LinNeg_N22 TSP2 Blood
9## ... ... ...
10## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC TSP10 Blood
11## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG TSP10 Blood
12## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG TSP10 Blood
13## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG TSP10 Blood
14## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG TSP10 Blood
15## anatomical_position method
16## <factor> <factor>
17## LinNeg_G15 NA smartseq
18## LinNeg_L11 NA smartseq
19## LinNeg_J16 NA smartseq
20## LinNeg_F5 NA smartseq
21## LinNeg_N22 NA smartseq
22## ... ... ...
23## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC NA 10X
24## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG NA 10X
25## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG NA 10X
26## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG NA 10X
27## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG NA 10X
28## cdna_plate library_plate
29## <factor> <factor>
30## LinNeg_G15 B113459 B133094
31## LinNeg_L11 B113459 B133094
32## LinNeg_J16 B113459 B133094
33## LinNeg_F5 B113459 B133094
34## LinNeg_N22 B113459 B133094
35## ... ... ...
36## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC nan nan
37## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG nan nan
38## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG nan nan
39## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG nan nan
40## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG nan nan
41## notes cdna_well
42## <factor> <factor>
43## LinNeg_G15 ImmuneLineageNeg G15
44## LinNeg_L11 ImmuneLineageNeg L11
45## LinNeg_J16 ImmuneLineageNeg J16
46## LinNeg_F5 ImmuneLineageNeg F5
47## LinNeg_N22 ImmuneLineageNeg N22
48## ... ... ...
49## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC Enriched nan
50## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG Enriched nan
51## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG Enriched nan
52## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG Enriched nan
53## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG Enriched nan
54## assay_ontology_term_id
55## <factor>
56## LinNeg_G15 EFO:0008931
57## LinNeg_L11 EFO:0008931
58## LinNeg_J16 EFO:0008931
59## LinNeg_F5 EFO:0008931
60## LinNeg_N22 EFO:0008931
61## ... ...
62## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC EFO:0009922
63## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG EFO:0009922
64## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG EFO:0009922
65## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG EFO:0009922
66## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG EFO:0009922
67## sample_id
68## <factor>
69## LinNeg_G15 TSP2_Blood_NA_SS2_B113459_B133094_ImmuneLineageNeg
70## LinNeg_L11 TSP2_Blood_NA_SS2_B113459_B133094_ImmuneLineageNeg
71## LinNeg_J16 TSP2_Blood_NA_SS2_B113459_B133094_ImmuneLineageNeg
72## LinNeg_F5 TSP2_Blood_NA_SS2_B113459_B133094_ImmuneLineageNeg
73## LinNeg_N22 TSP2_Blood_NA_SS2_B113459_B133094_ImmuneLineageNeg
74## ... ...
75## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC TSP10_Blood_NA_10X_1_1_Enriched
76## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG TSP10_Blood_NA_10X_1_1_Enriched
77## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG TSP10_Blood_NA_10X_1_1_Enriched
78## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG TSP10_Blood_NA_10X_1_1_Enriched
79## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG TSP10_Blood_NA_10X_1_1_Enriched
80## replicate
81## <integer>
82## LinNeg_G15 1
83## LinNeg_L11 1
84## LinNeg_J16 1
85## LinNeg_F5 1
86## LinNeg_N22 1
87## ... ...
88## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC 1
89## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG 1
90## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG 1
91## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG 1
92## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG 1
93## X10X_run
94## <factor>
95## LinNeg_G15 nan
96## LinNeg_L11 nan
97## LinNeg_J16 nan
98## LinNeg_F5 nan
99## LinNeg_N22 nan
100## ... ...
101## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC TSP10_Blood_NA_10X_1_1_Enriched
102## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG TSP10_Blood_NA_10X_1_1_Enriched
103## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG TSP10_Blood_NA_10X_1_1_Enriched
104## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG TSP10_Blood_NA_10X_1_1_Enriched
105## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG TSP10_Blood_NA_10X_1_1_Enriched
106## ambient_removal donor_method
107## <factor> <factor>
108## LinNeg_G15 None TSP2_smartseq
109## LinNeg_L11 None TSP2_smartseq
110## LinNeg_J16 None TSP2_smartseq
111## LinNeg_F5 None TSP2_smartseq
112## LinNeg_N22 None TSP2_smartseq
113## ... ... ...
114## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC decontx TSP10_10X
115## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG decontx TSP10_10X
116## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG decontx TSP10_10X
117## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG decontx TSP10_10X
118## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG decontx TSP10_10X
119## donor_assay
120## <factor>
121## LinNeg_G15 TSP2_SS2
122## LinNeg_L11 TSP2_SS2
123## LinNeg_J16 TSP2_SS2
124## LinNeg_F5 TSP2_SS2
125## LinNeg_N22 TSP2_SS2
126## ... ...
127## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC TSP10_10X_3Prime_v3.1
128## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG TSP10_10X_3Prime_v3.1
129## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG TSP10_10X_3Prime_v3.1
130## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG TSP10_10X_3Prime_v3.1
131## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG TSP10_10X_3Prime_v3.1
132## donor_tissue
133## <factor>
134## LinNeg_G15 TSP2_Blood
135## LinNeg_L11 TSP2_Blood
136## LinNeg_J16 TSP2_Blood
137## LinNeg_F5 TSP2_Blood
138## LinNeg_N22 TSP2_Blood
139## ... ...
140## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC TSP10_Blood
141## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG TSP10_Blood
142## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG TSP10_Blood
143## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG TSP10_Blood
144## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG TSP10_Blood
145## donor_tissue_assay
146## <factor>
147## LinNeg_G15 TSP2_Blood_SS2
148## LinNeg_L11 TSP2_Blood_SS2
149## LinNeg_J16 TSP2_Blood_SS2
150## LinNeg_F5 TSP2_Blood_SS2
151## LinNeg_N22 TSP2_Blood_SS2
152## ... ...
153## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC TSP10_Blood_10X_3Prime_v3.1
154## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG TSP10_Blood_10X_3Prime_v3.1
155## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG TSP10_Blood_10X_3Prime_v3.1
156## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG TSP10_Blood_10X_3Prime_v3.1
157## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG TSP10_Blood_10X_3Prime_v3.1
158## cell_type_ontology_term_id
159## <factor>
160## LinNeg_G15 CL:0000576
161## LinNeg_L11 CL:0000814
162## LinNeg_J16 CL:0000233
163## LinNeg_F5 CL:0000233
164## LinNeg_N22 CL:0000233
165## ... ...
166## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC CL:0000860
167## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG CL:0000236
168## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG CL:0000786
169## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG CL:0000625
170## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG CL:0000236
171## compartment
172## <factor>
173## LinNeg_G15 Immune
174## LinNeg_L11 Immune
175## LinNeg_J16 Immune
176## LinNeg_F5 Immune
177## LinNeg_N22 Immune
178## ... ...
179## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC Immune
180## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG Immune
181## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG Immune
182## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG Immune
183## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG Immune
184## broad_cell_class
185## <factor>
186## LinNeg_G15 myeloid leukocyte
187## LinNeg_L11 t cell
188## LinNeg_J16 hematopoietic cell
189## LinNeg_F5 hematopoietic cell
190## LinNeg_N22 hematopoietic cell
191## ... ...
192## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC myeloid leukocyte
193## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG lymphocyte of b lineage
194## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG lymphocyte of b lineage
195## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG t cell
196## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG lymphocyte of b lineage
197## free_annotation
198## <factor>
199## LinNeg_G15 monocyte
200## LinNeg_L11 type i nk t cell
201## LinNeg_J16 platelet
202## LinNeg_F5 platelet
203## LinNeg_N22 platelet
204## ... ...
205## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC classical monocyte
206## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG naive b cell
207## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG plasma cell
208## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG natural killer cell
209## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG naive b cell
210## manually_annotated
211## <factor>
212## LinNeg_G15 True
213## LinNeg_L11 False
214## LinNeg_J16 False
215## LinNeg_F5 False
216## LinNeg_N22 False
217## ... ...
218## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC True
219## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG True
220## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG False
221## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG True
222## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG True
223## published_2022
224## <factor>
225## LinNeg_G15 True
226## LinNeg_L11 False
227## LinNeg_J16 True
228## LinNeg_F5 True
229## LinNeg_N22 True
230## ... ...
231## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC True
232## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG True
233## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG False
234## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG True
235## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG True
236## nFeaturess_RNA_by_counts
237## <numeric>
238## LinNeg_G15 1674
239## LinNeg_L11 952
240## LinNeg_J16 915
241## LinNeg_F5 989
242## LinNeg_N22 1886
243## ... ...
244## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC 4605
245## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG 3156
246## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG 2278
247## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG 3409
248## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG 2323
249## total_counts total_counts_mt
250## <numeric> <numeric>
251## LinNeg_G15 193134 67101
252## LinNeg_L11 31754 1305
253## LinNeg_J16 3460323 115928
254## LinNeg_F5 3178758 100320
255## LinNeg_N22 6440280 64622
256## ... ... ...
257## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC 17180 188
258## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG 10264 358
259## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG 6652 1691
260## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG 8862 306
261## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG 6577 354
262## pct_counts_mt
263## <numeric>
264## LinNeg_G15 34.74324
265## LinNeg_L11 4.10972
266## LinNeg_J16 3.35021
267## LinNeg_F5 3.15595
268## LinNeg_N22 1.00340
269## ... ...
270## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC 1.09430
271## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG 3.48792
272## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG 25.42093
273## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG 3.45295
274## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG 5.38239
275## total_counts_ercc
276## <numeric>
277## LinNeg_G15 33428
278## LinNeg_L11 698
279## LinNeg_J16 134747
280## LinNeg_F5 69866
281## LinNeg_N22 84613
282## ... ...
283## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC 0
284## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG 0
285## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG 0
286## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG 0
287## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG 0
288## pct_counts_ercc X_scvi_batch
289## <numeric> <factor>
290## LinNeg_G15 17.30819 4
291## LinNeg_L11 2.19815 4
292## LinNeg_J16 3.89406 4
293## LinNeg_F5 2.19790 4
294## LinNeg_N22 1.31381 4
295## ... ... ...
296## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC 0 8
297## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG 0 8
298## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG 0 8
299## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG 0 8
300## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG 0 8
301## X_scvi_labels
302## <integer>
303## LinNeg_G15 0
304## LinNeg_L11 0
305## LinNeg_J16 0
306## LinNeg_F5 0
307## LinNeg_N22 0
308## ... ...
309## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC 0
310## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG 0
311## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG 0
312## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG 0
313## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG 0
314## scvi_leiden_donorassay_full
315## <factor>
316## LinNeg_G15 33
317## LinNeg_L11 33
318## LinNeg_J16 34
319## LinNeg_F5 34
320## LinNeg_N22 34
321## ... ...
322## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC 8
323## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG 2
324## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG 15
325## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG 16
326## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG 14
327## ethnicity_original
328## <factor>
329## LinNeg_G15 Black
330## LinNeg_L11 Black
331## LinNeg_J16 Black
332## LinNeg_F5 Black
333## LinNeg_N22 Black
334## ... ...
335## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC Hispanic
336## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG Hispanic
337## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG Hispanic
338## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG Hispanic
339## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG Hispanic
340## scvi_leiden_res05_tissue
341## <factor>
342## LinNeg_G15 15
343## LinNeg_L11 21
344## LinNeg_J16 29
345## LinNeg_F5 29
346## LinNeg_N22 29
347## ... ...
348## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC 2
349## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG 8
350## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG 22
351## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG 1
352## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG 8
353## sample_number
354## <integer>
355## LinNeg_G15 1
356## LinNeg_L11 1
357## LinNeg_J16 1
358## LinNeg_F5 1
359## LinNeg_N22 1
360## ... ...
361## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC 1
362## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG 1
363## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG 1
364## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG 1
365## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG 1
366## organism_ontology_term_id
367## <factor>
368## LinNeg_G15 NCBITaxon:9606
369## LinNeg_L11 NCBITaxon:9606
370## LinNeg_J16 NCBITaxon:9606
371## LinNeg_F5 NCBITaxon:9606
372## LinNeg_N22 NCBITaxon:9606
373## ... ...
374## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC NCBITaxon:9606
375## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG NCBITaxon:9606
376## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG NCBITaxon:9606
377## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG NCBITaxon:9606
378## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG NCBITaxon:9606
379## suspension_type tissue_type
380## <factor> <factor>
381## LinNeg_G15 cell tissue
382## LinNeg_L11 cell tissue
383## LinNeg_J16 cell tissue
384## LinNeg_F5 cell tissue
385## LinNeg_N22 cell tissue
386## ... ... ...
387## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC cell tissue
388## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG cell tissue
389## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG cell tissue
390## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG cell tissue
391## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG cell tissue
392## disease_ontology_term_id
393## <factor>
394## LinNeg_G15 PATO:0000461
395## LinNeg_L11 PATO:0000461
396## LinNeg_J16 PATO:0000461
397## LinNeg_F5 PATO:0000461
398## LinNeg_N22 PATO:0000461
399## ... ...
400## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC PATO:0000461
401## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG PATO:0000461
402## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG PATO:0000461
403## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG PATO:0000461
404## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG PATO:0000461
405## is_primary_data
406## <logical>
407## LinNeg_G15 FALSE
408## LinNeg_L11 FALSE
409## LinNeg_J16 FALSE
410## LinNeg_F5 FALSE
411## LinNeg_N22 FALSE
412## ... ...
413## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC FALSE
414## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG FALSE
415## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG FALSE
416## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG FALSE
417## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG FALSE
418## tissue_ontology_term_id
419## <factor>
420## LinNeg_G15 UBERON:0000178
421## LinNeg_L11 UBERON:0000178
422## LinNeg_J16 UBERON:0000178
423## LinNeg_F5 UBERON:0000178
424## LinNeg_N22 UBERON:0000178
425## ... ...
426## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC UBERON:0000178
427## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG UBERON:0000178
428## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG UBERON:0000178
429## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG UBERON:0000178
430## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG UBERON:0000178
431## sex_ontology_term_id
432## <factor>
433## LinNeg_G15 PATO:0000383
434## LinNeg_L11 PATO:0000383
435## LinNeg_J16 PATO:0000383
436## LinNeg_F5 PATO:0000383
437## LinNeg_N22 PATO:0000383
438## ... ...
439## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC PATO:0000384
440## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG PATO:0000384
441## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG PATO:0000384
442## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG PATO:0000384
443## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG PATO:0000384
444## self_reported_ethnicity_ontology_term_id
445## <factor>
446## LinNeg_G15 HANCESTRO:0016
447## LinNeg_L11 HANCESTRO:0016
448## LinNeg_J16 HANCESTRO:0016
449## LinNeg_F5 HANCESTRO:0016
450## LinNeg_N22 HANCESTRO:0016
451## ... ...
452## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC HANCESTRO:0014
453## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG HANCESTRO:0014
454## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG HANCESTRO:0014
455## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG HANCESTRO:0014
456## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG HANCESTRO:0014
457## development_stage_ontology_term_id
458## <factor>
459## LinNeg_G15 HsapDv:0000155
460## LinNeg_L11 HsapDv:0000155
461## LinNeg_J16 HsapDv:0000155
462## LinNeg_F5 HsapDv:0000155
463## LinNeg_N22 HsapDv:0000155
464## ... ...
465## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC HsapDv:0000127
466## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG HsapDv:0000127
467## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG HsapDv:0000127
468## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG HsapDv:0000127
469## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG HsapDv:0000127
470## cell_type
471## <factor>
472## LinNeg_G15 monocyte
473## LinNeg_L11 mature NK T cell
474## LinNeg_J16 platelet
475## LinNeg_F5 platelet
476## LinNeg_N22 platelet
477## ... ...
478## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC classical monocyte
479## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG B cell
480## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG plasma cell
481## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG CD8-positive, alpha-beta T cell
482## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG B cell
483## assay disease
484## <factor> <factor>
485## LinNeg_G15 Smart-seq2 normal
486## LinNeg_L11 Smart-seq2 normal
487## LinNeg_J16 Smart-seq2 normal
488## LinNeg_F5 Smart-seq2 normal
489## LinNeg_N22 Smart-seq2 normal
490## ... ... ...
491## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC 10x 3' v3 normal
492## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG 10x 3' v3 normal
493## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG 10x 3' v3 normal
494## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG 10x 3' v3 normal
495## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG 10x 3' v3 normal
496## organism sex tissue
497## <factor> <factor> <factor>
498## LinNeg_G15 Homo sapiens female blood
499## LinNeg_L11 Homo sapiens female blood
500## LinNeg_J16 Homo sapiens female blood
501## LinNeg_F5 Homo sapiens female blood
502## LinNeg_N22 Homo sapiens female blood
503## ... ... ... ...
504## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC Homo sapiens male blood
505## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG Homo sapiens male blood
506## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG Homo sapiens male blood
507## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG Homo sapiens male blood
508## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG Homo sapiens male blood
509## self_reported_ethnicity
510## <factor>
511## LinNeg_G15 African American or Afro-Caribbean
512## LinNeg_L11 African American or Afro-Caribbean
513## LinNeg_J16 African American or Afro-Caribbean
514## LinNeg_F5 African American or Afro-Caribbean
515## LinNeg_N22 African American or Afro-Caribbean
516## ... ...
517## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC Hispanic or Latin American
518## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG Hispanic or Latin American
519## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG Hispanic or Latin American
520## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG Hispanic or Latin American
521## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG Hispanic or Latin American
522## development_stage
523## <factor>
524## LinNeg_G15 61-year-old stage
525## LinNeg_L11 61-year-old stage
526## LinNeg_J16 61-year-old stage
527## LinNeg_F5 61-year-old stage
528## LinNeg_N22 61-year-old stage
529## ... ...
530## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC 33-year-old stage
531## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG 33-year-old stage
532## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG 33-year-old stage
533## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG 33-year-old stage
534## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG 33-year-old stage
535## observation_joinid nCount_RNA
536## <character> <numeric>
537## LinNeg_G15 q6>I#9TjHI 159702
538## LinNeg_L11 2rk&X7=xWu 31056
539## LinNeg_J16 IcM6sWp_ie 3325490
540## LinNeg_F5 #u#|Y$R>`- 3108881
541## LinNeg_N22 DExtx2aabf 6355661
542## ... ... ...
543## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC ZC+o|FE%%j 17178
544## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG IeBscd)Vb{ 10264
545## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG jNGtM=p@xV 6651
546## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG I*c09CPhkL 8861
547## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG fRD2Ce@4#! 6577
548## nFeature_RNA ident
549## <integer> <factor>
550## LinNeg_G15 1662 local
551## LinNeg_L11 946 local
552## LinNeg_J16 903 local
553## LinNeg_F5 975 local
554## LinNeg_N22 1872 local
555## ... ... ...
556## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGAGGTCGGTGTC 4603 local
557## TSP10_Blood_NA_10X_1_1_Enriched_TTTGGTTAGTACTGGG 3156 local
558## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGAGGGAGGTG 2277 local
559## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG 3408 local
560## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG 2323 local
we can extract just a part using subsetting
1colData(sce)[1:5,1:4]
1## DataFrame with 5 rows and 4 columns
2## donor_id tissue_in_publication anatomical_position method
3## <factor> <factor> <factor> <factor>
4## LinNeg_G15 TSP2 Blood NA smartseq
5## LinNeg_L11 TSP2 Blood NA smartseq
6## LinNeg_J16 TSP2 Blood NA smartseq
7## LinNeg_F5 TSP2 Blood NA smartseq
8## LinNeg_N22 TSP2 Blood NA smartseq
We can also extract data by column name directly from the sce object
1head(sce$donor_id)
1## [1] TSP2 TSP2 TSP2 TSP2 TSP2 TSP2
2## Levels: TSP1 TSP2 TSP7 TSP8 TSP10 TSP14 TSP21 TSP25 TSP27
and add new data
1random_group_labels <- sample(x = c("g1", "g2"), size = ncol(sce), replace = TRUE)
2sce$groups <- random_group_labels
let's check it
1colData(sce)[1:5, c("broad_cell_class", "cell_type", "groups")]
1## DataFrame with 5 rows and 3 columns
2## broad_cell_class cell_type groups
3## <factor> <factor> <character>
4## LinNeg_G15 myeloid leukocyte monocyte g2
5## LinNeg_L11 t cell mature NK T cell g2
6## LinNeg_J16 hematopoietic cell platelet g1
7## LinNeg_F5 hematopoietic cell platelet g1
8## LinNeg_N22 hematopoietic cell platelet g1
Gene metadata
Gene metadata can be accessed with the rowData
1head( rowData(sce) )
1## DataFrame with 6 rows and 0 columns
However, the conversion fails to import it. We will add it manually.
1rowData(sce) <- seurat[['RNA']][[]]
2head( rowData(sce) )
1## DataFrame with 6 rows and 16 columns
2## ensembl_id genome mt ercc
3## <character> <factor> <logical> <logical>
4## ENSG00000000003 ENSG00000000003.15 Gencode_v41 FALSE FALSE
5## ENSG00000000005 ENSG00000000005.6 Gencode_v41 FALSE FALSE
6## ENSG00000000419 ENSG00000000419.14 Gencode_v41 FALSE FALSE
7## ENSG00000000457 ENSG00000000457.14 Gencode_v41 FALSE FALSE
8## ENSG00000000460 ENSG00000000460.17 Gencode_v41 FALSE FALSE
9## ENSG00000000938 ENSG00000000938.13 Gencode_v41 FALSE FALSE
10## n_cells_by_counts mean_counts pct_dropout_by_counts
11## <numeric> <numeric> <numeric>
12## ENSG00000000003 161872 2.379617 91.8004
13## ENSG00000000005 9323 0.220273 99.5277
14## ENSG00000000419 461590 3.523875 76.6182
15## ENSG00000000457 156149 0.493041 92.0903
16## ENSG00000000460 120250 0.281519 93.9087
17## ENSG00000000938 255570 2.316143 87.0541
18## total_counts mean std feature_is_filtered
19## <numeric> <numeric> <numeric> <logical>
20## ENSG00000000003 4697694 6.24022e-04 0.02221597 FALSE
21## ENSG00000000005 434850 6.38723e-05 0.00764354 FALSE
22## ENSG00000000419 6956619 1.95665e-01 0.41830183 FALSE
23## ENSG00000000457 973332 7.00306e-02 0.26345956 FALSE
24## ENSG00000000460 555757 5.49991e-02 0.23468105 FALSE
25## ENSG00000000938 4572389 8.92788e-01 0.86412839 FALSE
26## feature_name feature_reference feature_biotype feature_length
27## <factor> <factor> <factor> <factor>
28## ENSG00000000003 TSPAN6 NCBITaxon:9606 gene 2396
29## ENSG00000000005 TNMD NCBITaxon:9606 gene 873
30## ENSG00000000419 DPM1 NCBITaxon:9606 gene 1262
31## ENSG00000000457 SCYL3 NCBITaxon:9606 gene 2916
32## ENSG00000000460 C1orf112 NCBITaxon:9606 gene 2661
33## ENSG00000000938 FGR NCBITaxon:9606 gene 2021
34## feature_type
35## <factor>
36## ENSG00000000003 protein_coding
37## ENSG00000000005 protein_coding
38## ENSG00000000419 protein_coding
39## ENSG00000000457 protein_coding
40## ENSG00000000460 protein_coding
41## ENSG00000000938 protein_coding
we can manipulate it as we have done with the colData.
1random_gene_data <- sample(x = c("g1", "g2"), size = nrow(sce), replace = TRUE)
2rowData(sce)$random <- random_gene_data
3rowData(sce)[1:3, c("random", "ensembl_id", "genome")]
1## DataFrame with 3 rows and 3 columns
2## random ensembl_id genome
3## <character> <character> <factor>
4## ENSG00000000003 g2 ENSG00000000003.15 Gencode_v41
5## ENSG00000000005 g2 ENSG00000000005.6 Gencode_v41
6## ENSG00000000419 g1 ENSG00000000419.14 Gencode_v41
Of note, there is a rowRanges slot in the sce object to keep genomic coordinates as GRanges or GRangesList. It stores the chromosome, start, and end coordinates of the features (genes for scRNAseq experiments or genomic regions for scATAC-seq for example) that can be accessed with rowRanges(sce) and manipulated using GenomicRanges package.
1rowRanges(sce)
1## GRangesList object of length 61759:
2## $ENSG00000000003
3## GRanges object with 0 ranges and 0 metadata columns:
4## seqnames ranges strand
5## <Rle> <IRanges> <Rle>
6## -------
7## seqinfo: no sequences
8##
9## $ENSG00000000005
10## GRanges object with 0 ranges and 0 metadata columns:
11## seqnames ranges strand
12## <Rle> <IRanges> <Rle>
13## -------
14## seqinfo: no sequences
15##
16## $ENSG00000000419
17## GRanges object with 0 ranges and 0 metadata columns:
18## seqnames ranges strand
19## <Rle> <IRanges> <Rle>
20## -------
21## seqinfo: no sequences
22##
23## ...
24## <61756 more elements>
Assays
Assays store the primary data of the object such as a matrix of counts, rows correspond to features (genes) and columns correspond to samples (cells). Thus, SCE assays would be equivalent to Seurat's layers.
SCE has several predefined assays:
counts: Raw count data.normcounts: Normalized values on the same scale as the original counts. For example, counts divided by cell-specific size factors that are centred at unity.logcounts: Log-transformed counts, e.g. using log base 2 and a pseudo-count of 1.cpm: Counts-per-million normalized data.tpm: Transcripts-per-million normalized data.
The most used are counts and logcounts. We can access these assays with dedicated functions, counts and logcounts.
1counts(sce)[1:4,1:5]
1## 4 x 5 sparse Matrix of class "dgCMatrix"
2## LinNeg_G15 LinNeg_L11 LinNeg_J16 LinNeg_F5 LinNeg_N22
3## ENSG00000000003 . . . . .
4## ENSG00000000005 . . . . .
5## ENSG00000000419 . . . . .
6## ENSG00000000457 . . . . .
1logcounts(sce)[45:49,20:25]
1## 5 x 6 sparse Matrix of class "dgCMatrix"
2## LinNeg_P20 LinNeg_J10 LinNeg_M15 LinNeg_C23 LinNeg_E13
3## ENSG00000003989 . . . . .
4## ENSG00000004059 . 0.005375588 0.95498431 . .
5## ENSG00000004139 . . . . .
6## ENSG00000004142 . . 0.07273076 . .
7## ENSG00000004399 . 0.047370646 . . .
8## LinNeg_K21
9## ENSG00000003989 .
10## ENSG00000004059 1.1101098
11## ENSG00000004139 .
12## ENSG00000004142 .
13## ENSG00000004399 0.1566256
The general method assay(sce, <Assay-name>) can also be used to access or set predefined or custom assays.
1assay(sce, 'my.assay') <- counts(sce)
2assay(sce, 'my.assay')[1:4,1:5]
1## 4 x 5 sparse Matrix of class "dgCMatrix"
2## LinNeg_G15 LinNeg_L11 LinNeg_J16 LinNeg_F5 LinNeg_N22
3## ENSG00000000003 . . . . .
4## ENSG00000000005 . . . . .
5## ENSG00000000419 . . . . .
6## ENSG00000000457 . . . . .
Dimensional reduction
Dimensional reductions can be accessed with reducedDims.
1reducedDims(sce)
1## List of length 7
2## names(7): PCA SCVI ... UMAP_TISSUE_SCVI_DONORASSAY UNCORRECTED_UMAP
Let's check the PCA.
1dim(reducedDim(sce, "PCA"))
1## [1] 85233 50
It contains 50 components for the cells of the object. Let's inspect the first two components.
1reducedDim(sce, "PCA")[1:5,1:2]
1## PC_1 PC_2
2## LinNeg_G15 8.216231 27.61160
3## LinNeg_L11 5.790802 32.35962
4## LinNeg_J16 6.373532 25.54363
5## LinNeg_F5 7.180281 24.49034
6## LinNeg_N22 7.337627 21.94072
In addition to the PC coordinate matrix, the object contains three attributes:
percentVar: Percentage of variance explained by each PC. This may not sum to 100 if not all PCs are reported.varExplained: The variance explained by each PC.rotation: The loadings for all genes in each PC.
1attr(reducedDim(sce, 'PCA'), "percentVar")
1## NULL
1attr(reducedDim(sce, 'PCA'), "varExplained")
1## NULL
1attr(reducedDim(sce, 'PCA'), "rotation")
1## NULL
They were not exported :( but we can added if we wish
1attr(reducedDim(sce, 'PCA'), "percentVar") <- seq( from= 10, to= 0, by = -0.5 )
Alternative Experiments
SCE uses alternative experiments, or altExps, to store data for a different set of features of the same cells (colnames). The tipical use for this would be multimodal single-cell omics experiments such as CITE-seq or ATAC-seq data for example. With altExps we can combine SCE objects for 'other' omics data for downstream use but separated from the assays in the main experiment holding the RNA-seq counts.
Let's see an example.
1altExp( sce, 'ADT' ) <- ADT.sce # Antigen derived tags from CITEseq experiment
2sce
1## class: SingleCellExperiment
2## dim: 61759 85233
3## metadata(0):
4## assays(3): counts logcounts my.assay
5## rownames(61759): ENSG00000000003 ENSG00000000005 ... ENSG00000290165
6## ENSG00000290166
7## rowData names(17): ensembl_id genome ... feature_type random
8## colnames(85233): LinNeg_G15 LinNeg_L11 ...
9## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG
10## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG
11## colData names(57): donor_id tissue_in_publication ... ident groups
12## reducedDimNames(7): PCA SCVI ... UMAP_TISSUE_SCVI_DONORASSAY
13## UNCORRECTED_UMAP
14## mainExpName: RNA
15## altExpNames(1): ADT
explore or use the ADT (Antigen derived tags) data as follows
1altExp( sce, 'ADT' )
1## class: SingleCellExperiment
2## dim: 4 85233
3## metadata(0):
4## assays(1): counts
5## rownames(4): CD3 CD4 CD8a CD14
6## rowData names(2): antibody method
7## colnames(85233): LinNeg_G15 LinNeg_L11 ...
8## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCAAGCCG
9## TSP10_Blood_NA_10X_1_1_Enriched_TTTGTTGTCCGCGATG
10## colData names(6): donor_id tissue_in_publication ... free_annotation
11## cell_type
12## reducedDimNames(0):
13## mainExpName: NULL
14## altExpNames(0):
Thus, SCE alternative experiments would correspond to Seurat's assays, I know a little bit confusing...
Save you work
As with the Seurat object, you can save the SCE object to a rds file.
1saveRDS( sce, 'blood_cells.rds')
Useful tips
Manual object creation
To create an SCE object from scratch, you just need to read the expression data, metadata and dimensional reductions separately and then create a new object.
Prepare the data
1counts <- read.csv( file.path( data.path, 'counts.csv' ), row.names = 1 ) # raw counts
2logcounts <- read.csv( file.path( data.path, 'logcounts.csv' ), row.names = 1 ) # normalized counts
3cell_metadata <- read.csv( file.path( data.path, 'cell_metadata.csv' ), row.names = 1 )
4gene_metadata <- read.csv( file.path( data.path, 'gene_metadata.csv' ), row.names = 1 )
5PCA <- read.csv( file.path( data.path, 'PCA.csv' ), row.names = 1 )
create the object
1manual.sce <- SingleCellExperiment(
2 assays=list(counts=as.matrix(counts),
3 logcounts=as.matrix(logcounts)
4 ),
5 colData = cell_metadata,
6 rowData = gene_metadata,
7 reducedDims=SimpleList(PCA=as.matrix(PCA))
8)
Check the object
1manual.sce
1## class: SingleCellExperiment
2## dim: 30157 3879
3## metadata(0):
4## assays(2): counts logcounts
5## rownames(30157): ENSG00000238009 ENSG00000241860 ... ENSG00000288057
6## ENSG00000228786
7## rowData names(3): GeneID GeneName Chromosome
8## colnames(3879): CELL2_N4 CELL20_N2 ... CELL74524_N1 CELL74525_N1
9## colData names(16): cell sample ... scDblFinder.score scDblFinder.class
10## reducedDimNames(1): PCA
11## mainExpName: NULL
12## altExpNames(0):
Big datasets
As happened with Scanpy or Seurat, some experiments are too big and need to be handled differently. Using the HDF5Array package will allow us to create an object without loading the whole matrix on memory.
We need first to create an HDF5 array file to store the counts matrix, for the example we will use the same counts matrix we used in the manual SCE creation.
1library(HDF5Array)
2hdf5_file <- file.path(data.path, "large_counts.h5")
3hdf5_counts <- writeHDF5Array(counts, filepath = hdf5_file, name = "counts")
After that we just need to create an object as usual.
1h5.sce <- SingleCellExperiment(
2 assays = list(counts = hdf5_counts), # HDF5-backed counts matrix
3 rowData = gene_metadata,
4 colData = cell_metadata
5)
Take home messages
- The SingleCellExperiment package default Bioconductor's package to store single cell data.
- Unlike AnnData and Scanpy, columns correspond to the cells’ barcodes and rows are the gene IDs.
- It can be directly used with over 70 single-cell-related Bioconductor packages.
- SCE can easily store millions of cells if we use it with HDF5Array.
Next posts of this series will cover how to perform single cell data analysis with SingleCellExperiment.
If you liked this post, your can visit my previous posts on Seurat and Scanpy post, if you haven't yet.
Further reading
- Orchestrating Single-Cell Analysis with Bioconductor – book: https://bioconductor.org/books/release/OSCA/
- Orchestrating Large-Scale Single-Cell Analysis with Bioconductor - ISMB: https://bioconductor.github.io/ISMB.OSCA/