ENACT Visium HD cell segmentation and annotation
Spatial transcriptomics reached a whole new level with the latest release from 10x Genomics, Visium HD. This state-of-the-art technology offers exceptional spatial resolution in transcriptomics, enabling researchers to map gene expression with unprecedented detail. From tumor microenvironments to intricate tissue architecture, Visium HD is transforming cellular biology experiments--- pixel by pixel.
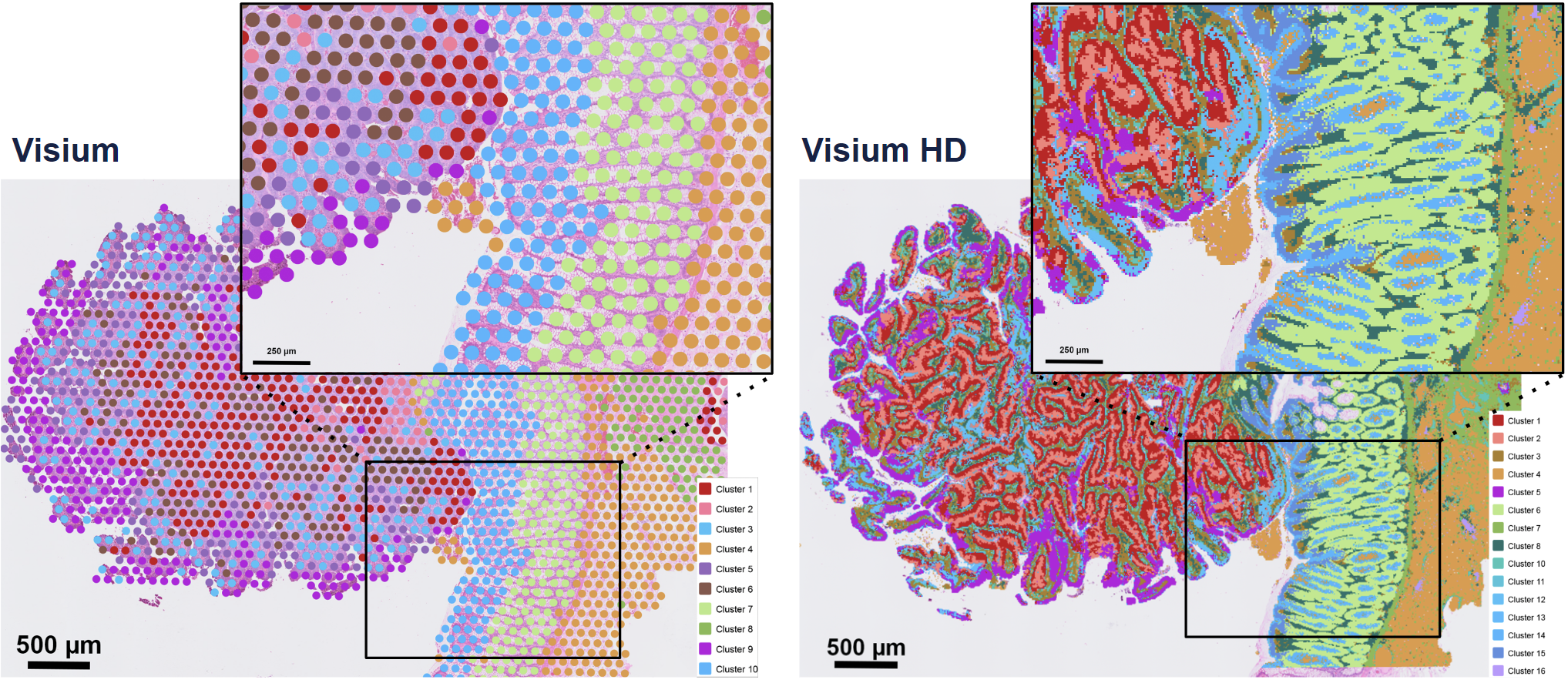
The key advantage of Visium HD over Visium is moving from discrete spots to a continuous lawn of oligonucleotides arrayed into millions of 2 x 2 µm barcoded squares that captures whole transcriptome expression.
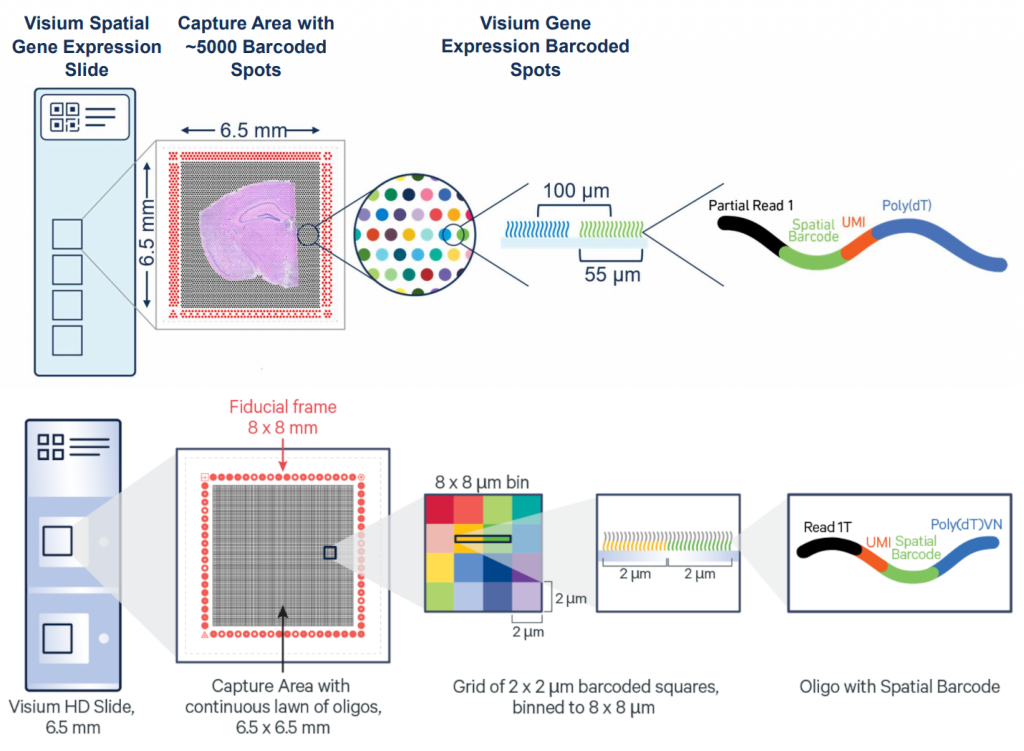
Eventhough Visium HD provides extreme resolution, we still miss a key part to truly understand the biology of the system: single-cell individuality. Luckily for us, we can use segmentation techniques to reconstructs cells from the high resolution Visium HD data.
Edit. Nov 2025
Space Ranger v4.0 (released in June 2025) now automatically performs nucleus-based cell segmentation for Visium HD and Visium HD 3' data if H&E images are provided.
Following my previous posts in VisiumHD nuclei segmentation and Visium HD cell segmentation with Bin2Cell, today we test ENACT, a complete end-to-end pipeline that includes Visium HD Cell Segmentation and Cell type annotation. Recently published in Bioinformatcis, ENACT also builds on the StarDist, the object detection technique used in the nuclei segmentation post.
You will need TensorFlow to perform this analysis. I run my code with jupyter notebook from a docker image as explained in my Using TensorFlow in 2025 post.
To make it work I had to run docker as follows:
1docker run -p 8888:8888 -v ${PWD}:/tf/visium --gpus all --rm tensorflow/tensorflow:2.15.0-gpu-jupyter
and install the python packages using the terminal from jupyter.
Then, we can install ENACT with a single pip command.
1pip install enact-SO
As discussed in my TensorFlow post, the last versions of TensorFlow do not want to work in my computer so I had to manually install the version 2.15.0.
1pip install --force-reinstall tensorflow==2.15.0
Once ENACT is installed we just need to download the tmap_template into a folder named templates next to the jupyter notebook file, and the config.yaml file to the folder specified later in configs_path
Modify the configs file to the specific setting of your experiment, see here for detailed guidelines.
The following parameters configure analysis name and ENACT's Required Files: output path (cache_dir), and input paths for the visium HD data
1analysis_name: <analysis-name>
2cache_dir: <path-to-store-enact-outputs>
3paths:
4 wsi_path: <path-to-whole-slide-image>
5 visiumhd_h5_path: <path-to-counts-file>
6 tissue_positions_path: <path-to-tissue-positions>
next define ENACT's core parameters
1params:
2 bin_to_cell_method: "weighted_by_cluster"
3 cell_annotation_method: "celltypist"
4 cell_typist_model: "Adult_Mouse_Gut.pkl"
bin-to-cell_method can be "naive", "weighted_by_area", "weighted_by_gene" or "weighted_by_cluster".
cell_annotation_method can be "cellassign", "celltypist" or "sargent" (if installed).
cell_typist_model is the model weights for the cell annotation step, select the appropriate on from the official website
ENACT allows users to define the distance for nuclei expansion to obtain cell boundaries. ENACT starts by segmenting nuclei using Stardist, followed by generating Voronoi polygons based on nuclei centroids. Nuclei boundaries are then expanded within their respective Voronoi polygons using the user-defined distance to ensure non-overlapping cell boundaries. The following part of the configs file controls this.
1 nucleus_expansion: True # Flag to enable nuclei expansion to get cell boundaries
2 expand_by_nbins: 2 # Number of bins to expand the nuclei by to get cell boundaries
Run ENACT
As mentioned above, we need the templates folder to be in the same directory as the jupyter notebook. I also downloaded the configs file here.
1!ls
1templates
2Intestine.ENACT.ipynb
3configs.yaml
load packages
1from enact.pipeline import ENACT
2import yaml
read the configs file that was previously downloaded
1configs_path = "configs.yaml"
2with open(configs_path, "r") as stream:
3 configs = yaml.safe_load(stream)
4so_hd = ENACT(configs_dict=configs)
12025-03-23 01:01:29,442 - ENACT - INFO - <initiate_instance_variables> ENACT running with the following configurations:
2 analysis_name: Intestine.ENACT
3 run_synthetic: False
4 cache_dir: /tf/visium/ENACT
5 wsi_path: /tf/visium/Visium_HD_Mouse_Small_Intestine_tissue_image.btf
6 visiumhd_h5_path: /tf/visium/binned_outputs/square_002um/filtered_feature_bc_matrix.h5
7 tissue_positions_path: /tf/visium/binned_outputs/square_002um/spatial/tissue_positions.parquet
8 segmentation: True
9 bin_to_geodataframes: True
10 bin_to_cell_assignment: True
11 cell_type_annotation: True
12 seg_method: stardist
13 image_type: he
14 nucleus_expansion: True
15 expand_by_nbins: 2
16 patch_size: 4000
17 bin_representation: polygon
18 bin_to_cell_method: weighted_by_area
19 cell_annotation_method: celltypist
20 cell_typist_model: Adult_Mouse_Gut.pkl
21 use_hvg: False
22 n_hvg: 1000
23 destripe_norm: False
24 n_clusters: 4
25 n_pcs: 250
26 chunks_to_run: []
27 block_size: 4096
28 prob_thresh: 0.005
29 overlap_thresh: 0.001
30 min_overlap: 128
31 context: 128
32 n_tiles: (8,8,1)
33 stardist_modelname: 2D_versatile_he
34 channel_to_segment: 2
35 cell_markers: {'Enterocytes': ['Cbr1', 'Plin2', 'Gls', 'Plin3', 'Dab1', 'Pmepa1', 'Acsl5', 'Hmox1', 'Abcg2', 'Cd36'], 'Goblet cells': ['Manf', 'Krt7', 'Ccl9', 'Muc13', 'Phgr1', 'Cdx2', 'Aqp3', 'Creb3L1', 'Guca2A', 'Klk1'], 'Enteroendocrine cells': ['Fabp5', 'Cpe', 'Enpp2', 'Chgb', 'Alcam', 'Chga', 'Pax6', 'Neurod1', 'Cck', 'Isl1'], 'Paneth cells': ['Gpx2', 'Fabp4', 'Lyz1', 'Kcnn4', 'Lgals2', 'Guca2B', 'Lgr4', 'Defa24', 'Il4Ra', 'Guca2A'], 'Crypt cells': ['Prom1', 'Hopx', 'Msi1', 'Olfm4', 'Kcne3', 'Bmi1', 'Axin2', 'Kcnq1', 'Ascl2', 'Lrig1'], 'Smooth muscle cells': ['Bgn', 'Myl9', 'Pcp4L1', 'Itga1', 'Nrp2', 'Mylk', 'Ehd2', 'Fabp4', 'Acta2', 'Ogn'], 'B cells': ['Cd52', 'Bcl11A', 'Ebf1', 'Cd74', 'Ptprc', 'Pold4', 'Ighm', 'Cd14', 'Creld2', 'Fli1'], 'T cells': ['Cd81', 'Junb', 'Cd52', 'Ptprcap', 'H2-Q7', 'Ccl6', 'Bcl2', 'Maff', 'Ccl4', 'Ccl3'], 'NK cells': ['Ctla2A', 'Ccl4', 'Cd3G', 'Ccl3', 'Nkg7', 'Lat', 'Dusp2', 'Itgam', 'Fhl2', 'Ccl5']}
run ENACT with just one command
1so_hd.run_enact()
12025-03-23 01:01:42,159 - ENACT - INFO - <load_image> Successfully loaded image!
2<load_image> Successfully loaded image!
32025-03-23 01:02:05,780 - ENACT - INFO - <normalize_image> Successfully normalized image!
4<normalize_image> Successfully normalized image!
5
6
7Found model '2D_versatile_he' for 'StarDist2D'.
8Loading network weights from 'weights_best.h5'.
9Loading thresholds from 'thresholds.json'.
10Using default values: prob_thresh=0.692478, nms_thresh=0.3.
11effective: block_size=(4096, 4096, 3), min_overlap=(128, 128, 0), context=(128, 128, 0)
12
13
14100%|██████████| 42/42 [08:02<00:00, 11.50s/it]
152025-03-23 01:10:09,155 - ENACT - INFO - <run_segmentation> Successfully segmented cells!
16<run_segmentation> Successfully segmented cells!
172025-03-23 01:10:49,122 - ENACT - INFO - <convert_stardist_output_to_gdf> Mean nuclei area: 177.57601532126913
18<convert_stardist_output_to_gdf> Mean nuclei area: 177.57601532126913
192025-03-23 01:10:49,160 - ENACT - INFO - <run_enact> Expanding nuclei to get cell boundaries
20<run_enact> Expanding nuclei to get cell boundaries
212025-03-23 01:10:50,063 - ENACT - INFO - <get_bin_size> Bin size computed: 7.303833516704913 pixels
22<get_bin_size> Bin size computed: 7.303833516704913 pixels
232025-03-23 01:12:14,481 - ENACT - INFO - <expand_nuclei_with_voronoi> Number of unexpanded cells: 30
24<expand_nuclei_with_voronoi> Number of unexpanded cells: 30
252025-03-23 01:12:14,903 - ENACT - INFO - <convert_stardist_output_to_gdf> Mean nuclei area after expansion: 577.3306682089849
26<convert_stardist_output_to_gdf> Mean nuclei area after expansion: 577.3306682089849
272025-03-23 01:12:57,935 - ENACT - INFO - <split_df_to_chunks> Splitting into chunks. output_dir: /tf/visium/ENACT/Intestine.ENACT/chunks/cells_gdf
28<split_df_to_chunks> Splitting into chunks. output_dir: /tf/visium/ENACT/Intestine.ENACT/chunks/cells_gdf
29100%|██████████| 34/34 [00:10<00:00, 3.38it/s]
302025-03-23 01:13:35,568 - ENACT - INFO - <load_visiumhd_dataset> Missing the following markers: {'Cd3G', 'Guca2B', 'Il4Ra', 'Ctla2A', 'Guca2A', 'Defa24', 'Bcl11A', 'Creb3L1', 'Pcp4L1'}
31<load_visiumhd_dataset> Missing the following markers: {'Cd3G', 'Guca2B', 'Il4Ra', 'Ctla2A', 'Guca2A', 'Defa24', 'Bcl11A', 'Creb3L1', 'Pcp4L1'}
322025-03-23 01:13:36,474 - ENACT - INFO - <generate_bin_polys> Generating bin polygons. num_bins: 5479660
33<generate_bin_polys> Generating bin polygons. num_bins: 5479660
34100%|██████████| 5479660/5479660 [01:44<00:00, 52367.00it/s]
352025-03-23 01:15:27,721 - ENACT - INFO - <split_df_to_chunks> Splitting into chunks. output_dir: /tf/visium/ENACT/Intestine.ENACT/chunks/bins_gdf
36<split_df_to_chunks> Splitting into chunks. output_dir: /tf/visium/ENACT/Intestine.ENACT/chunks/bins_gdf
37100%|██████████| 37/37 [01:35<00:00, 2.59s/it]
382025-03-23 01:17:30,754 - ENACT - INFO - <load_visiumhd_dataset> Missing the following markers: {'Cd3G', 'Guca2B', 'Il4Ra', 'Ctla2A', 'Guca2A', 'Defa24', 'Bcl11A', 'Creb3L1', 'Pcp4L1'}
39<load_visiumhd_dataset> Missing the following markers: {'Cd3G', 'Guca2B', 'Il4Ra', 'Ctla2A', 'Guca2A', 'Defa24', 'Bcl11A', 'Creb3L1', 'Pcp4L1'}
402025-03-23 01:17:31,515 - ENACT - INFO - <assign_bins_to_cells> Assigning bins to cells using weighted_by_area method
41<assign_bins_to_cells> Assigning bins to cells using weighted_by_area method
42<assign_bins_to_cells> Processed patch_1_3.csv using weighted_by_area. Mean count per cell: 9.827049428195927
43100%|██████████| 34/34 [15:23<00:00, 27.16s/it]
442025-03-23 01:32:55,095 - ENACT - INFO - <assign_bins_to_cells> Successfully assigned bins to cells!
45<assign_bins_to_cells> Successfully assigned bins to cells!
462025-03-23 01:32:55,248 - ENACT - INFO - <initiate_instance_variables> ENACT running with the following configurations:
47 analysis_name: Intestine.ENACT
48 run_synthetic: False
49 cache_dir: /tf/visium/ENACT
50 wsi_path: /tf/visium/Visium_HD_Mouse_Small_Intestine_tissue_image.btf
51 visiumhd_h5_path: /tf/visium/binned_outputs/square_002um/filtered_feature_bc_matrix.h5
52 tissue_positions_path: /tf/visium/binned_outputs/square_002um/spatial/tissue_positions.parquet
53 segmentation: True
54 bin_to_geodataframes: True
55 bin_to_cell_assignment: True
56 cell_type_annotation: True
57 seg_method: stardist
58 image_type: he
59 nucleus_expansion: True
60 expand_by_nbins: 2
61 patch_size: 4000
62 bin_representation: polygon
63 bin_to_cell_method: weighted_by_area
64 cell_annotation_method: celltypist
65 cell_typist_model: Adult_Mouse_Gut.pkl
66 use_hvg: True
67 n_hvg: 1000
68 destripe_norm: False
69 n_clusters: 4
70 n_pcs: 250
71 chunks_to_run: []
72 block_size: 4096
73 prob_thresh: 0.005
74 overlap_thresh: 0.001
75 min_overlap: 128
76 context: 128
77 n_tiles: (8,8,1)
78 stardist_modelname: 2D_versatile_he
79 channel_to_segment: 2
80 cell_markers: {'Enterocytes': ['Cbr1', 'Plin2', 'Gls', 'Plin3', 'Dab1', 'Pmepa1', 'Acsl5', 'Hmox1', 'Abcg2', 'Cd36'], 'Goblet cells': ['Manf', 'Krt7', 'Ccl9', 'Muc13', 'Phgr1', 'Cdx2', 'Aqp3', 'Creb3L1', 'Guca2A', 'Klk1'], 'Enteroendocrine cells': ['Fabp5', 'Cpe', 'Enpp2', 'Chgb', 'Alcam', 'Chga', 'Pax6', 'Neurod1', 'Cck', 'Isl1'], 'Paneth cells': ['Gpx2', 'Fabp4', 'Lyz1', 'Kcnn4', 'Lgals2', 'Guca2B', 'Lgr4', 'Defa24', 'Il4Ra', 'Guca2A'], 'Crypt cells': ['Prom1', 'Hopx', 'Msi1', 'Olfm4', 'Kcne3', 'Bmi1', 'Axin2', 'Kcnq1', 'Ascl2', 'Lrig1'], 'Smooth muscle cells': ['Bgn', 'Myl9', 'Pcp4L1', 'Itga1', 'Nrp2', 'Mylk', 'Ehd2', 'Fabp4', 'Acta2', 'Ogn'], 'B cells': ['Cd52', 'Bcl11A', 'Ebf1', 'Cd74', 'Ptprc', 'Pold4', 'Ighm', 'Cd14', 'Creld2', 'Fli1'], 'T cells': ['Cd81', 'Junb', 'Cd52', 'Ptprcap', 'H2-Q7', 'Ccl6', 'Bcl2', 'Maff', 'Ccl4', 'Ccl3'], 'NK cells': ['Ctla2A', 'Ccl4', 'Cd3G', 'Ccl3', 'Nkg7', 'Lat', 'Dusp2', 'Itgam', 'Fhl2', 'Ccl5']}
81
82<initiate_instance_variables> ENACT running with the following configurations:
83 analysis_name: Intestine.ENACT
84 run_synthetic: False
85 cache_dir: /tf/visium/ENACT
86 wsi_path: /tf/visium/Visium_HD_Mouse_Small_Intestine_tissue_image.btf
87 visiumhd_h5_path: /tf/visium/binned_outputs/square_002um/filtered_feature_bc_matrix.h5
88 tissue_positions_path: /tf/visium/binned_outputs/square_002um/spatial/tissue_positions.parquet
89 segmentation: True
90 bin_to_geodataframes: True
91 bin_to_cell_assignment: True
92 cell_type_annotation: True
93 seg_method: stardist
94 image_type: he
95 nucleus_expansion: True
96 expand_by_nbins: 2
97 patch_size: 4000
98 bin_representation: polygon
99 bin_to_cell_method: weighted_by_area
100 cell_annotation_method: celltypist
101 cell_typist_model: Adult_Mouse_Gut.pkl
102 use_hvg: True
103 n_hvg: 1000
104 destripe_norm: False
105 n_clusters: 4
106 n_pcs: 250
107 chunks_to_run: []
108 block_size: 4096
109 prob_thresh: 0.005
110 overlap_thresh: 0.001
111 min_overlap: 128
112 context: 128
113 n_tiles: (8,8,1)
114 stardist_modelname: 2D_versatile_he
115 channel_to_segment: 2
116 cell_markers: {'Enterocytes': ['Cbr1', 'Plin2', 'Gls', 'Plin3', 'Dab1', 'Pmepa1', 'Acsl5', 'Hmox1', 'Abcg2', 'Cd36'], 'Goblet cells': ['Manf', 'Krt7', 'Ccl9', 'Muc13', 'Phgr1', 'Cdx2', 'Aqp3', 'Creb3L1', 'Guca2A', 'Klk1'], 'Enteroendocrine cells': ['Fabp5', 'Cpe', 'Enpp2', 'Chgb', 'Alcam', 'Chga', 'Pax6', 'Neurod1', 'Cck', 'Isl1'], 'Paneth cells': ['Gpx2', 'Fabp4', 'Lyz1', 'Kcnn4', 'Lgals2', 'Guca2B', 'Lgr4', 'Defa24', 'Il4Ra', 'Guca2A'], 'Crypt cells': ['Prom1', 'Hopx', 'Msi1', 'Olfm4', 'Kcne3', 'Bmi1', 'Axin2', 'Kcnq1', 'Ascl2', 'Lrig1'], 'Smooth muscle cells': ['Bgn', 'Myl9', 'Pcp4L1', 'Itga1', 'Nrp2', 'Mylk', 'Ehd2', 'Fabp4', 'Acta2', 'Ogn'], 'B cells': ['Cd52', 'Bcl11A', 'Ebf1', 'Cd74', 'Ptprc', 'Pold4', 'Ighm', 'Cd14', 'Creld2', 'Fli1'], 'T cells': ['Cd81', 'Junb', 'Cd52', 'Ptprcap', 'H2-Q7', 'Ccl6', 'Bcl2', 'Maff', 'Ccl4', 'Ccl3'], 'NK cells': ['Ctla2A', 'Ccl4', 'Cd3G', 'Ccl3', 'Nkg7', 'Lat', 'Dusp2', 'Itgam', 'Fhl2', 'Ccl5']}
117
118📂 Storing models in /root/.celltypist/data/models
119💾 Total models to download: 1
120⏩ Skipping [1/1]: Adult_Mouse_Gut.pkl (file exists)
121🔬 Input data has 393713 cells and 1073 genes
122🔗 Matching reference genes in the model
123🧬 301 features used for prediction
124⚖️ Scaling input data
125🖋️ Predicting labels
126✅ Prediction done!
127classifier.py (126): DataFrame is highly fragmented. This is usually the result of calling `frame.insert` many times, which has poor performance. Consider joining all columns at once using pd.concat(axis=1) instead. To get a de-fragmented frame, use `newframe = frame.copy()`
1282025-03-23 01:33:32,326 - ENACT - INFO - <run_cell_type_annotation> Successfully ran CellTypist on Data.
129<run_cell_type_annotation> Successfully ran CellTypist on Data.
1302025-03-23 01:33:32,328 - ENACT - INFO - <initiate_instance_variables> ENACT running with the following configurations:
131 analysis_name: Intestine.ENACT
132 run_synthetic: False
133 cache_dir: /tf/visium/ENACT
134 wsi_path: /tf/visium/Visium_HD_Mouse_Small_Intestine_tissue_image.btf
135 visiumhd_h5_path: /tf/visium/binned_outputs/square_002um/filtered_feature_bc_matrix.h5
136 tissue_positions_path: /tf/visium/binned_outputs/square_002um/spatial/tissue_positions.parquet
137 segmentation: True
138 bin_to_geodataframes: True
139 bin_to_cell_assignment: True
140 cell_type_annotation: True
141 seg_method: stardist
142 image_type: he
143 nucleus_expansion: True
144 expand_by_nbins: 2
145 patch_size: 4000
146 bin_representation: polygon
147 bin_to_cell_method: weighted_by_area
148 cell_annotation_method: celltypist
149 cell_typist_model: Adult_Mouse_Gut.pkl
150 use_hvg: True
151 n_hvg: 1000
152 destripe_norm: False
153 n_clusters: 4
154 n_pcs: 250
155 chunks_to_run: []
156 block_size: 4096
157 prob_thresh: 0.005
158 overlap_thresh: 0.001
159 min_overlap: 128
160 context: 128
161 n_tiles: (8,8,1)
162 stardist_modelname: 2D_versatile_he
163 channel_to_segment: 2
164 cell_markers: {'Enterocytes': ['Cbr1', 'Plin2', 'Gls', 'Plin3', 'Dab1', 'Pmepa1', 'Acsl5', 'Hmox1', 'Abcg2', 'Cd36'], 'Goblet cells': ['Manf', 'Krt7', 'Ccl9', 'Muc13', 'Phgr1', 'Cdx2', 'Aqp3', 'Creb3L1', 'Guca2A', 'Klk1'], 'Enteroendocrine cells': ['Fabp5', 'Cpe', 'Enpp2', 'Chgb', 'Alcam', 'Chga', 'Pax6', 'Neurod1', 'Cck', 'Isl1'], 'Paneth cells': ['Gpx2', 'Fabp4', 'Lyz1', 'Kcnn4', 'Lgals2', 'Guca2B', 'Lgr4', 'Defa24', 'Il4Ra', 'Guca2A'], 'Crypt cells': ['Prom1', 'Hopx', 'Msi1', 'Olfm4', 'Kcne3', 'Bmi1', 'Axin2', 'Kcnq1', 'Ascl2', 'Lrig1'], 'Smooth muscle cells': ['Bgn', 'Myl9', 'Pcp4L1', 'Itga1', 'Nrp2', 'Mylk', 'Ehd2', 'Fabp4', 'Acta2', 'Ogn'], 'B cells': ['Cd52', 'Bcl11A', 'Ebf1', 'Cd74', 'Ptprc', 'Pold4', 'Ighm', 'Cd14', 'Creld2', 'Fli1'], 'T cells': ['Cd81', 'Junb', 'Cd52', 'Ptprcap', 'H2-Q7', 'Ccl6', 'Bcl2', 'Maff', 'Ccl4', 'Ccl3'], 'NK cells': ['Ctla2A', 'Ccl4', 'Cd3G', 'Ccl3', 'Nkg7', 'Lat', 'Dusp2', 'Itgam', 'Fhl2', 'Ccl5']}
165
166<initiate_instance_variables> ENACT running with the following configurations:
167 analysis_name: Intestine.ENACT
168 run_synthetic: False
169 cache_dir: /tf/visium/ENACT
170 wsi_path: /tf/visium/Visium_HD_Mouse_Small_Intestine_tissue_image.btf
171 visiumhd_h5_path: /tf/visium/binned_outputs/square_002um/filtered_feature_bc_matrix.h5
172 tissue_positions_path: /tf/visium/binned_outputs/square_002um/spatial/tissue_positions.parquet
173 segmentation: True
174 bin_to_geodataframes: True
175 bin_to_cell_assignment: True
176 cell_type_annotation: True
177 seg_method: stardist
178 image_type: he
179 nucleus_expansion: True
180 expand_by_nbins: 2
181 patch_size: 4000
182 bin_representation: polygon
183 bin_to_cell_method: weighted_by_area
184 cell_annotation_method: celltypist
185 cell_typist_model: Adult_Mouse_Gut.pkl
186 use_hvg: True
187 n_hvg: 1000
188 destripe_norm: False
189 n_clusters: 4
190 n_pcs: 250
191 chunks_to_run: []
192 block_size: 4096
193 prob_thresh: 0.005
194 overlap_thresh: 0.001
195 min_overlap: 128
196 context: 128
197 n_tiles: (8,8,1)
198 stardist_modelname: 2D_versatile_he
199 channel_to_segment: 2
200 cell_markers: {'Enterocytes': ['Cbr1', 'Plin2', 'Gls', 'Plin3', 'Dab1', 'Pmepa1', 'Acsl5', 'Hmox1', 'Abcg2', 'Cd36'], 'Goblet cells': ['Manf', 'Krt7', 'Ccl9', 'Muc13', 'Phgr1', 'Cdx2', 'Aqp3', 'Creb3L1', 'Guca2A', 'Klk1'], 'Enteroendocrine cells': ['Fabp5', 'Cpe', 'Enpp2', 'Chgb', 'Alcam', 'Chga', 'Pax6', 'Neurod1', 'Cck', 'Isl1'], 'Paneth cells': ['Gpx2', 'Fabp4', 'Lyz1', 'Kcnn4', 'Lgals2', 'Guca2B', 'Lgr4', 'Defa24', 'Il4Ra', 'Guca2A'], 'Crypt cells': ['Prom1', 'Hopx', 'Msi1', 'Olfm4', 'Kcne3', 'Bmi1', 'Axin2', 'Kcnq1', 'Ascl2', 'Lrig1'], 'Smooth muscle cells': ['Bgn', 'Myl9', 'Pcp4L1', 'Itga1', 'Nrp2', 'Mylk', 'Ehd2', 'Fabp4', 'Acta2', 'Ogn'], 'B cells': ['Cd52', 'Bcl11A', 'Ebf1', 'Cd74', 'Ptprc', 'Pold4', 'Ighm', 'Cd14', 'Creld2', 'Fli1'], 'T cells': ['Cd81', 'Junb', 'Cd52', 'Ptprcap', 'H2-Q7', 'Ccl6', 'Bcl2', 'Maff', 'Ccl4', 'Ccl3'], 'NK cells': ['Ctla2A', 'Ccl4', 'Cd3G', 'Ccl3', 'Nkg7', 'Lat', 'Dusp2', 'Itgam', 'Fhl2', 'Ccl5']}
201
202... storing 'patch_id' as categorical
2032025-03-23 01:34:09,298 - ENACT - INFO - <load_image> Successfully loaded image!
204<load_image> Successfully loaded image!
2052025-03-23 01:34:17,235 - ENACT - INFO - <package_results> Packaged CellTypist results
206<package_results> Packaged CellTypist results
207
208
209
210 Sample ready to visualize on TissUUmaps. To install TissUUmaps, follow the instructions at:
211
212 https://tissuumaps.github.io/TissUUmaps-docs/docs/intro/installation.html#.
213
214 To view the the sample, follow the instructions at:
215
216 https://tissuumaps.github.io/TissUUmaps-docs/docs/starting/projects.html#loading-projects
217
218 TissUUmaps project file is located here:
219
220 /tf/visium/ENACT/Intestine.ENACT/tmap/weighted_by_area|celltypist_tmap.tmap
221
And that's it! Cell segmentation and annotation are done. You can find here the explanation of the output folder structure. For the rest of the analysis, we will use the annotated annData object.
Exploring ENACT results
Load annotated annData object.
1import anndata as ad
2import scanpy as sc
3file_path = "ENACT_output/Intestine.ENACT/chunks/weighted_by_area/celltypist_results/cells_adata.h5"
4adata = sc.read_h5ad(file_path)
We start with a quick filtering to keep genes expressed in at least 3 cells and cells with at least 1 count.
1sc.pp.filter_genes(adata, min_cells=3)
2sc.pp.filter_cells(adata, min_counts=1)
and follow with expression normalization using scanpy
1sc.pp.normalize_total(adata, inplace=True)
2sc.pp.log1p(adata)
Marker Genes
As in previous post, we will use specific cell type marker genes to evaluate the results.
- Paneth cell: Lyz1
- Goblet cell: Muc2
- Enterocyte cell: Fabp2
- Plasma cell: Jchain
Since we already have cell types, we will use them instead of calculating clusters to group the cells.
1markers = { 'Paneth': 'Lyz1', 'Goblet': 'Muc2', 'Plasma Cell': 'Jchain', 'Enterocyte': 'Fabp2' }
2p = sc.pl.dotplot(adata, markers, groupby='cell_type', dendrogram=True, return_fig=True)
3p.add_totals().style(dot_edge_color='black', dot_edge_lw=0.5).show()
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
Cell In[12], line 2
1 markers = { 'Paneth': 'Lyz1', 'Goblet': 'Muc2', 'Plasma Cell': 'Jchain', 'Enterocyte': 'Fabp2' }
----> 2 p = sc.pl.dotplot(adata, markers, groupby='cell_type', dendrogram=True, return_fig=True)
3 p.add_totals().style(dot_edge_color='black', dot_edge_lw=0.5).show()
KeyError: "Could not find keys '['Fabp2', 'Jchain', 'Muc2']' in columns of `adata.obs` or in adata.var_names."
Surprisingly, most of the markers are not found in the data. Let's check the expression of the marker present in the data.
1markers = ['Lyz1']
2sc.pl.dotplot(adata, markers, groupby='cell_type', dendrogram=True)
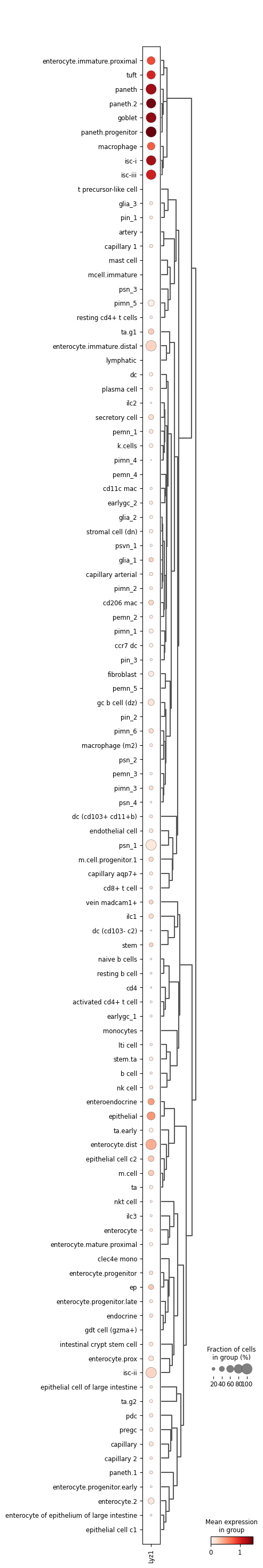
Paneth.progenitor and paneth.2 cells are the ones with highest expression of Lyz1. Other cell types with high expression include paneth, goblet and isc-i.
Next we explore the annData object.
1adata
AnnData object with n_obs × n_vars = 383968 × 1072
obs: 'cell_type', 'patch_id', 'n_counts'
var: 'n_cells'
obsm: 'spatial', 'stats'
1adata.X.shape
(383968, 1072)
1adata.var
| Rgs20 |
|---|
| Adhfe1 |
| Kcnb2 |
| Jph1 |
| Khdc1c |
| ... |
| Vegfd |
| Asb11 |
| Arhgap6 |
| Amelx |
| mt-Nd5 |
1073 rows × 0 columns
We can clearly see the number of genes in the object is 1073.
It turns out there are a couple of settings in the config file that control the number of genes used for the analysis. The default is to use 1000 highly variable genes (HVG) plus the cell markers specified in the config file.
1 use_hvg: True
2 n_hvg: 1000
Sadly, when I tried to run the analysis setting use_hvg to False (n_hvg = 0 or 1000), the python kernel died. The same happened when I tried to run the analysis with use_hvg = True and n_hvg = 10000. Preventing a depper analysis of the results based on gene expression, as performed for the previous posts on this topic.
If you have been able to run ENACT with all the genes let me know in the comments.
Take home messages
- Visium HD offers unparalleled spatial resolution providing a level of detail not seen before
- ENACT allows to perform a whole analysis, from the visium results to cell type annotation, with a single command.
- ENACt also allow to run bin2cell destripe normalization if desired.
Further reading
- ENACT publication (Kamel et al., 2025)
- ENACT gihub
- CellTypist publication (Dominguez Conde et al., 2022)
- CellTypist github, by Bin2Cell authors