Autoencoders for Single Cell Data Analysis
We live in the age of AI, machine learning, deep learning and LLMs. Today we will focus in autoencoders.
Autoencoders can be defined as a class of unsupervised neural networks designed to learn compact, informative representations of data by encoding inputs into a low-dimensional latent space and then reconstructing them. I see autoencoders as the deep learning version of PCA.
1##
2## Attaching package: 'dplyr'
1## The following objects are masked from 'package:stats':
2##
3## filter, lag
1## The following objects are masked from 'package:base':
2##
3## intersect, setdiff, setequal, union
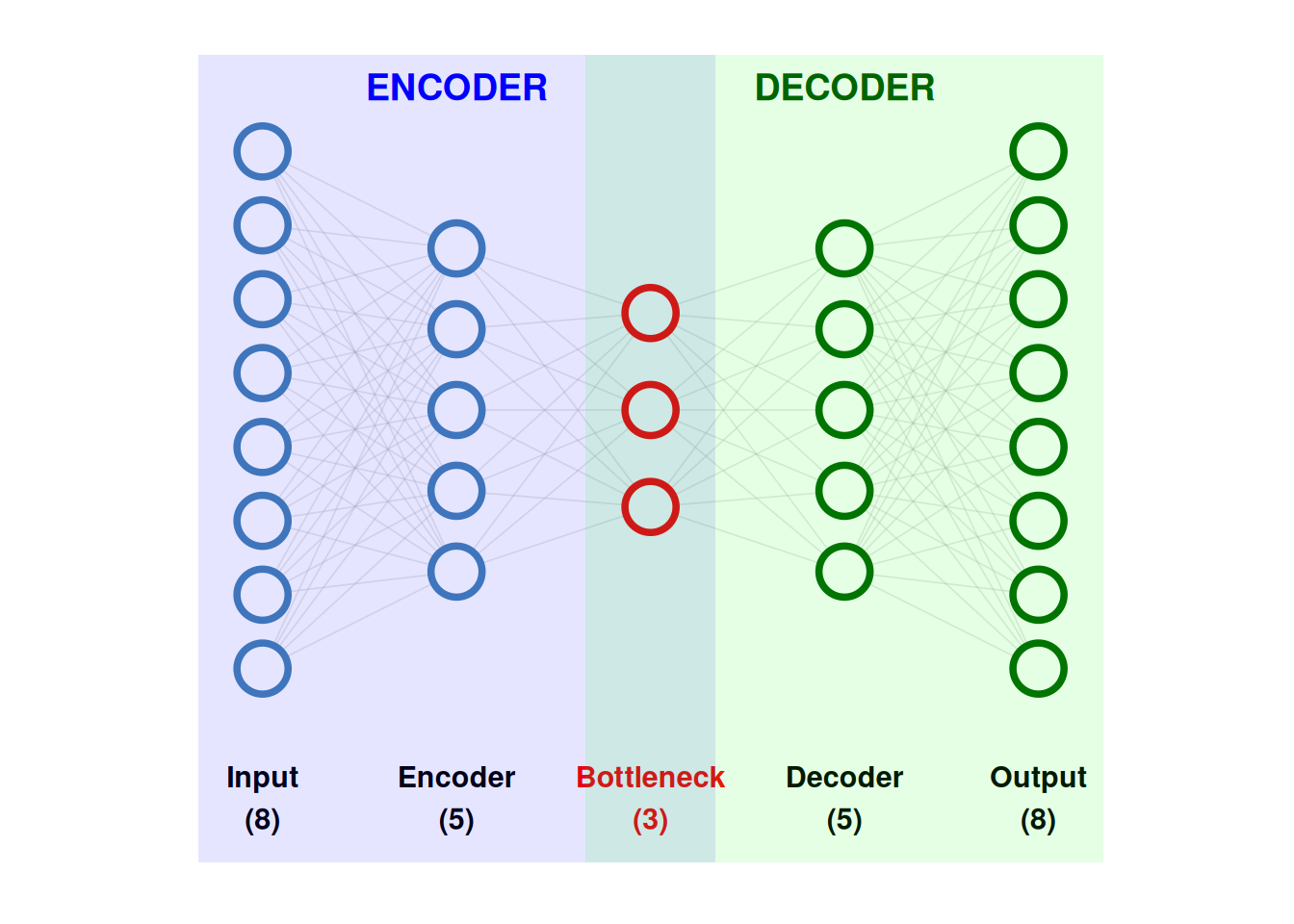
These are the basic components of an autoencoder:
- Input layer: Receives the original data and passes it to the network for encoding.
- Encoder: Transforms the input data into a lower-dimensional representation by learning informative features.
- Latent space (bottleneck): A compressed representation that captures the most salient information needed to reconstruct the input.
- Decoder: Maps the latent representation back to the original data space.
- Output (reconstruction): The network’s attempt to reproduce the original input as accurately as possible.
In single-cell, autoencoders can be used to model the high dimensionality, sparsity, and technical noise inherent in single-cell RNA-sequencing data. For example, scVI (single-cell variational inference) uses a variational autoencoder framework to perform batch correction, dimensionality reduction, and probabilistic modeling of gene expression, while methods like DCA (Deep Count Autoencoder) focus on denoising and handling dropout events, and SAUCIE uses autoencoders for visualization, clustering, and batch effect removal.
we will use the pancreas dataset as in my slingshot and RNA velocity post.
1library(zellkonverter)
1## Registered S3 method overwritten by 'zellkonverter':
2## method from
3## py_to_r.pandas.core.arrays.categorical.Categorical reticulate
1sce <- readH5AD( file.path( data.path, 'endocrinogenesis_day15.h5ad' ) )
1sce
1## class: SingleCellExperiment
2## dim: 27998 3696
3## metadata(5): clusters_coarse_colors clusters_colors day_colors
4## neighbors pca
5## assays(3): X spliced unspliced
6## rownames(27998): Xkr4 Gm37381 ... Gm20837 Erdr1
7## rowData names(1): highly_variable_genes
8## colnames(3696): AAACCTGAGAGGGATA AAACCTGAGCCTTGAT ... TTTGTCATCGAATGCT
9## TTTGTCATCTGTTTGT
10## colData names(4): clusters_coarse clusters S_score G2M_score
11## reducedDimNames(2): X_pca X_umap
12## mainExpName: NULL
13## altExpNames(0):
save X assay as logcounts to make our life easier.
1library(SingleCellExperiment)
1## Loading required package: SummarizedExperiment
1## Loading required package: MatrixGenerics
1## Loading required package: matrixStats
1##
2## Attaching package: 'matrixStats'
1## The following object is masked from 'package:dplyr':
2##
3## count
1##
2## Attaching package: 'MatrixGenerics'
1## The following objects are masked from 'package:matrixStats':
2##
3## colAlls, colAnyNAs, colAnys, colAvgsPerRowSet, colCollapse,
4## colCounts, colCummaxs, colCummins, colCumprods, colCumsums,
5## colDiffs, colIQRDiffs, colIQRs, colLogSumExps, colMadDiffs,
6## colMads, colMaxs, colMeans2, colMedians, colMins, colOrderStats,
7## colProds, colQuantiles, colRanges, colRanks, colSdDiffs, colSds,
8## colSums2, colTabulates, colVarDiffs, colVars, colWeightedMads,
9## colWeightedMeans, colWeightedMedians, colWeightedSds,
10## colWeightedVars, rowAlls, rowAnyNAs, rowAnys, rowAvgsPerColSet,
11## rowCollapse, rowCounts, rowCummaxs, rowCummins, rowCumprods,
12## rowCumsums, rowDiffs, rowIQRDiffs, rowIQRs, rowLogSumExps,
13## rowMadDiffs, rowMads, rowMaxs, rowMeans2, rowMedians, rowMins,
14## rowOrderStats, rowProds, rowQuantiles, rowRanges, rowRanks,
15## rowSdDiffs, rowSds, rowSums2, rowTabulates, rowVarDiffs, rowVars,
16## rowWeightedMads, rowWeightedMeans, rowWeightedMedians,
17## rowWeightedSds, rowWeightedVars
1## Loading required package: GenomicRanges
1## Loading required package: stats4
1## Loading required package: BiocGenerics
1##
2## Attaching package: 'BiocGenerics'
1## The following objects are masked from 'package:dplyr':
2##
3## combine, intersect, setdiff, union
1## The following objects are masked from 'package:stats':
2##
3## IQR, mad, sd, var, xtabs
1## The following objects are masked from 'package:base':
2##
3## anyDuplicated, aperm, append, as.data.frame, basename, cbind,
4## colnames, dirname, do.call, duplicated, eval, evalq, Filter, Find,
5## get, grep, grepl, intersect, is.unsorted, lapply, Map, mapply,
6## match, mget, order, paste, pmax, pmax.int, pmin, pmin.int,
7## Position, rank, rbind, Reduce, rownames, sapply, setdiff, table,
8## tapply, union, unique, unsplit, which.max, which.min
1## Loading required package: S4Vectors
1##
2## Attaching package: 'S4Vectors'
1## The following objects are masked from 'package:dplyr':
2##
3## first, rename
1## The following object is masked from 'package:utils':
2##
3## findMatches
1## The following objects are masked from 'package:base':
2##
3## expand.grid, I, unname
1## Loading required package: IRanges
1##
2## Attaching package: 'IRanges'
1## The following objects are masked from 'package:dplyr':
2##
3## collapse, desc, slice
1## Loading required package: GenomeInfoDb
1## Loading required package: Biobase
1## Welcome to Bioconductor
2##
3## Vignettes contain introductory material; view with
4## 'browseVignettes()'. To cite Bioconductor, see
5## 'citation("Biobase")', and for packages 'citation("pkgname")'.
1##
2## Attaching package: 'Biobase'
1## The following object is masked from 'package:MatrixGenerics':
2##
3## rowMedians
1## The following objects are masked from 'package:matrixStats':
2##
3## anyMissing, rowMedians
1logcounts( sce ) <- assay( sce, 'X' )
prep input data for neural network, to speed up computations we will work with the 2000 most variable genes.
1library(scran)
1## Loading required package: scuttle
1dec <- modelGeneVar(sce)
1## Warning in regularize.values(x, y, ties, missing(ties), na.rm = na.rm):
2## collapsing to unique 'x' values
1hvg.pbmc.var <- getTopHVGs(dec, n=2000)
2hvg.mat <- logcounts(sce)[hvg.pbmc.var,]
3write.csv( as.matrix(hvg.mat), 'hgv.genes.mat.csv' )
to use tensorflow, the easiest way is to run it with docker as explained in my post.
1docker run -p 8888:8888 -v ./:/tf/autoencoder --gpus all -it --rm tensorflow/tensorflow:2.15.0-gpu-jupyter
select new notebook and then run the following python code, we start importing tensorflow and required classes
1import tensorflow as tf
2from tensorflow.keras.layers import Input, Dense
3from tensorflow.keras.models import Model
12025-12-13 18:55:50.357811: E external/local_xla/xla/stream_executor/cuda/cuda_dnn.cc:9261] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered
22025-12-13 18:55:50.357833: E external/local_xla/xla/stream_executor/cuda/cuda_fft.cc:607] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered
32025-12-13 18:55:50.358524: E external/local_xla/xla/stream_executor/cuda/cuda_blas.cc:1515] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
42025-12-13 18:55:50.362622: I tensorflow/core/platform/cpu_feature_guard.cc:182] This TensorFlow binary is optimized to use available CPU instructions in performance-critical operations.
5To enable the following instructions: AVX2 FMA, in other operations, rebuild TensorFlow with the appropriate compiler flags.
now install the packages that are not included in this docker, if you are going to do this regularly just create a new image starting from this one.
1!pip install pandas numpy scikit-learn
1Requirement already satisfied: pandas in /usr/local/lib/python3.11/dist-packages (2.3.3)
2Requirement already satisfied: numpy in /usr/local/lib/python3.11/dist-packages (1.26.2)
3Requirement already satisfied: scikit-learn in /usr/local/lib/python3.11/dist-packages (1.8.0)
4Requirement already satisfied: python-dateutil>=2.8.2 in /usr/local/lib/python3.11/dist-packages (from pandas) (2.8.2)
5Requirement already satisfied: pytz>=2020.1 in /usr/local/lib/python3.11/dist-packages (from pandas) (2025.2)
6Requirement already satisfied: tzdata>=2022.7 in /usr/local/lib/python3.11/dist-packages (from pandas) (2025.3)
7Requirement already satisfied: scipy>=1.10.0 in /usr/local/lib/python3.11/dist-packages (from scikit-learn) (1.16.3)
8Requirement already satisfied: joblib>=1.3.0 in /usr/local/lib/python3.11/dist-packages (from scikit-learn) (1.5.2)
9Requirement already satisfied: threadpoolctl>=3.2.0 in /usr/local/lib/python3.11/dist-packages (from scikit-learn) (3.6.0)
10Requirement already satisfied: six>=1.5 in /usr/lib/python3/dist-packages (from python-dateutil>=2.8.2->pandas) (1.16.0)
11[33mWARNING: Running pip as the 'root' user can result in broken permissions and conflicting behaviour with the system package manager. It is recommended to use a virtual environment instead: https://pip.pypa.io/warnings/venv[0m[33m
12[0m
13[1m[[0m[34;49mnotice[0m[1;39;49m][0m[39;49m A new release of pip is available: [0m[31;49m23.3.1[0m[39;49m -> [0m[32;49m25.3[0m
14[1m[[0m[34;49mnotice[0m[1;39;49m][0m[39;49m To update, run: [0m[32;49mpython3 -m pip install --upgrade pip[0m
import the packages
1import pandas as pd
2import numpy as np
3from sklearn.preprocessing import StandardScaler
read the gene information
1csv_file_path = 'hgv.genes.mat.csv'
2data = pd.read_csv(csv_file_path, index_col=0)
take a look to the data
1data.iloc[:5, :5]
| AAACCTGAGAGGGATA | AAACCTGAGCCTTGAT | AAACCTGAGGCAATTA | AAACCTGCATCATCCC | AAACCTGGTAAGTGGC | |
|---|---|---|---|---|---|
| Pyy | 103 | 0 | 11 | 0 | 1 |
| Ghrl | 0 | 1 | 0 | 0 | 0 |
| Iapp | 0 | 0 | 27 | 0 | 0 |
| Gcg | 0 | 0 | 0 | 0 | 0 |
| Ins1 | 0 | 0 | 0 | 0 | 0 |
A common approach in 'regular' autoencoders is transforming the data with StandardScaler from sckitlearn to scale the data. However, StandardScaler is usually not recommended for scRNA-seq autoencoders.
- Log2 normalized data is is non-negative.
- Many zeros remain biologically meaningful.
- Variance across genes is informative
so we split input dat into train and test data. Instead of manual splitting, this approach shuffles the data avoiding posible Batch effects leakage and cell-type ordering bias
1from sklearn.model_selection import train_test_split
2train_data, test_data = train_test_split(input_data, test_size=0.2, random_state=42)
check the split by printing the shape (dimensions) of the resulting arrays
1print("Training data shape:", train_data.shape)
2print("Testing data shape:", test_data.shape)
1Training data shape: (2956, 2000)
2Testing data shape: (740, 2000)
It is time to configure our autoencoder. We start defining the number of neurons per layer
1neurons = [2**x for x in range(1,11)]
2neurons
1[2, 4, 8, 16, 32, 64, 128, 256, 512, 1024]
now the learning rate and the number of epochs.
The learning rate is a hyperparameter that controls how much a model's parameters are updated in response to the estimated error at each training step.
The number of epochs refers to how many complete passes the learning algorithm makes over the entire training dataset, with more epochs allowing the model to learn more.
1custom_learning_rate = 0.001
2epochs = 50
the next function will create the autoencoder
1def create_autoencoder(neurons, input_shape):
2 input_layer = Input(shape=(input_shape,))
3
4 # Encoder layers
5 encoder_output = input_layer
6 for i in range(len(neurons)-1, 0, -1):
7 encoder_output = Dense(neurons[i], activation='elu')(encoder_output)
8
9 # Hidden layer with a name
10 hidden_layer = Dense(neurons[0], activation='linear', name='hidden_layer')(encoder_output)
11
12 # Decoder layers
13 decoder_output = hidden_layer
14 for i in range(1, len(neurons)):
15 decoder_output = Dense(neurons[i], activation='elu')(decoder_output)
16
17 # Output layer
18 output_layer = Dense(input_shape, activation='linear')(decoder_output)
19 # output_layer = Dense(input_shape, activation='sigmoid')(decoder_output) # changed because input goes from -1 to 1
20
21 autoencoder = Model(inputs=input_layer, outputs=output_layer)
22 return autoencoder
now we use it to set up our autoencoder
1autoencoder = create_autoencoder(neurons, train_data.shape[1])
12025-12-13 19:04:48.696696: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
22025-12-13 19:04:48.703243: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
32025-12-13 19:04:48.706844: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
42025-12-13 19:04:48.709844: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
52025-12-13 19:04:48.711871: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
62025-12-13 19:04:48.713724: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
72025-12-13 19:04:48.834992: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
82025-12-13 19:04:48.835741: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
92025-12-13 19:04:48.836391: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
102025-12-13 19:04:48.837020: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1929] Created device /job:localhost/replica:0/task:0/device:GPU:0 with 712 MB memory: -> device: 0, name: NVIDIA GeForce RTX 2070 SUPER, pci bus id: 0000:01:00.0, compute capability: 7.5
prep the optimizer and compile the autoencoder
1optimizer = tf.keras.optimizers.Adam(learning_rate=custom_learning_rate)
2autoencoder.compile(optimizer=optimizer, loss='mean_squared_error')
show the model using summary
1autoencoder.summary()
1Model: "model"
2_________________________________________________________________
3 Layer (type) Output Shape Param #
4=================================================================
5 input_1 (InputLayer) [(None, 2000)] 0
6
7 dense (Dense) (None, 1024) 2049024
8
9 dense_1 (Dense) (None, 512) 524800
10
11 dense_2 (Dense) (None, 256) 131328
12
13 dense_3 (Dense) (None, 128) 32896
14
15 dense_4 (Dense) (None, 64) 8256
16
17 dense_5 (Dense) (None, 32) 2080
18
19 dense_6 (Dense) (None, 16) 528
20
21 dense_7 (Dense) (None, 8) 136
22
23 dense_8 (Dense) (None, 4) 36
24
25 hidden_layer (Dense) (None, 2) 10
26
27 dense_9 (Dense) (None, 4) 12
28
29 dense_10 (Dense) (None, 8) 40
30
31 dense_11 (Dense) (None, 16) 144
32
33 dense_12 (Dense) (None, 32) 544
34
35 dense_13 (Dense) (None, 64) 2112
36
37 dense_14 (Dense) (None, 128) 8320
38
39 dense_15 (Dense) (None, 256) 33024
40
41 dense_16 (Dense) (None, 512) 131584
42
43 dense_17 (Dense) (None, 1024) 525312
44
45 dense_18 (Dense) (None, 2000) 2050000
46
47=================================================================
48Total params: 5500186 (20.98 MB)
49Trainable params: 5500186 (20.98 MB)
50Non-trainable params: 0 (0.00 Byte)
51_________________________________________________________________
time to train the autoencoder using the training data
1H = autoencoder.fit(x = train_data, y = train_data,
2 validation_data=(test_data, test_data),
3 batch_size = 32, epochs = epochs, verbose = 1)
1Epoch 1/50
2
3
42025-12-13 19:05:16.520701: I external/local_xla/xla/service/service.cc:168] XLA service 0x7de6e060eb90 initialized for platform CUDA (this does not guarantee that XLA will be used). Devices:
52025-12-13 19:05:16.520721: I external/local_xla/xla/service/service.cc:176] StreamExecutor device (0): NVIDIA GeForce RTX 2070 SUPER, Compute Capability 7.5
62025-12-13 19:05:16.524636: I tensorflow/compiler/mlir/tensorflow/utils/dump_mlir_util.cc:269] disabling MLIR crash reproducer, set env var `MLIR_CRASH_REPRODUCER_DIRECTORY` to enable.
72025-12-13 19:05:16.537091: I external/local_xla/xla/stream_executor/cuda/cuda_dnn.cc:454] Loaded cuDNN version 8906
8WARNING: All log messages before absl::InitializeLog() is called are written to STDERR
9I0000 00:00:1765652716.584968 2328 device_compiler.h:186] Compiled cluster using XLA! This line is logged at most once for the lifetime of the process.
10
11
1293/93 [==============================] - 6s 18ms/step - loss: 32.5919 - val_loss: 31.4853
13Epoch 2/50
1493/93 [==============================] - 2s 16ms/step - loss: 23.7953 - val_loss: 28.6923
15Epoch 3/50
1693/93 [==============================] - 2s 17ms/step - loss: 23.2020 - val_loss: 35.4802
17Epoch 4/50
1893/93 [==============================] - 1s 15ms/step - loss: 22.8094 - val_loss: 27.7426
19Epoch 5/50
2093/93 [==============================] - 1s 14ms/step - loss: 25.9521 - val_loss: 30.0849
21Epoch 6/50
2293/93 [==============================] - 1s 16ms/step - loss: 23.6428 - val_loss: 28.0149
23Epoch 7/50
2493/93 [==============================] - 1s 13ms/step - loss: 21.9675 - val_loss: 26.9624
25Epoch 8/50
2693/93 [==============================] - 1s 15ms/step - loss: 21.5971 - val_loss: 26.5356
27Epoch 9/50
2893/93 [==============================] - 1s 16ms/step - loss: 21.0882 - val_loss: 25.2487
29Epoch 10/50
3093/93 [==============================] - 2s 17ms/step - loss: 20.9234 - val_loss: 24.0937
31Epoch 11/50
3293/93 [==============================] - 1s 15ms/step - loss: 20.4022 - val_loss: 24.3812
33Epoch 12/50
3493/93 [==============================] - 1s 15ms/step - loss: 20.0690 - val_loss: 24.9853
35Epoch 13/50
3693/93 [==============================] - 1s 13ms/step - loss: 19.7373 - val_loss: 24.5243
37Epoch 14/50
3893/93 [==============================] - 2s 17ms/step - loss: 23.2898 - val_loss: 22.5970
39Epoch 15/50
4093/93 [==============================] - 1s 16ms/step - loss: 23.6739 - val_loss: 31.1010
41Epoch 16/50
4293/93 [==============================] - 1s 14ms/step - loss: 25.1240 - val_loss: 28.3302
43Epoch 17/50
4493/93 [==============================] - 1s 16ms/step - loss: 25.6185 - val_loss: 30.2079
45Epoch 18/50
4693/93 [==============================] - 1s 13ms/step - loss: 22.9125 - val_loss: 27.1545
47Epoch 19/50
4893/93 [==============================] - 1s 12ms/step - loss: 20.8052 - val_loss: 24.0625
49Epoch 20/50
5093/93 [==============================] - 1s 15ms/step - loss: 18.9230 - val_loss: 21.5312
51Epoch 21/50
5293/93 [==============================] - 2s 18ms/step - loss: 17.6285 - val_loss: 19.5485
53Epoch 22/50
5493/93 [==============================] - 2s 17ms/step - loss: 17.4163 - val_loss: 19.8080
55Epoch 23/50
5693/93 [==============================] - 2s 18ms/step - loss: 19.2971 - val_loss: 22.0913
57Epoch 24/50
5893/93 [==============================] - 1s 11ms/step - loss: 27.8436 - val_loss: 31.5397
59Epoch 25/50
6093/93 [==============================] - 1s 13ms/step - loss: 22.9990 - val_loss: 24.4272
61Epoch 26/50
6293/93 [==============================] - 1s 10ms/step - loss: 21.5256 - val_loss: 22.1785
63Epoch 27/50
6493/93 [==============================] - 1s 16ms/step - loss: 19.2715 - val_loss: 19.6551
65Epoch 28/50
6693/93 [==============================] - 2s 17ms/step - loss: 18.4090 - val_loss: 21.0072
67Epoch 29/50
6893/93 [==============================] - 2s 17ms/step - loss: 17.2127 - val_loss: 18.2983
69Epoch 30/50
7093/93 [==============================] - 1s 12ms/step - loss: 19.0098 - val_loss: 20.4128
71Epoch 31/50
7293/93 [==============================] - 1s 13ms/step - loss: 23.4715 - val_loss: 25.3339
73Epoch 32/50
7493/93 [==============================] - 1s 14ms/step - loss: 19.5397 - val_loss: 20.0218
75Epoch 33/50
7693/93 [==============================] - 2s 17ms/step - loss: 16.9510 - val_loss: 15.9626
77Epoch 34/50
7893/93 [==============================] - 1s 14ms/step - loss: 17.7965 - val_loss: 21.5455
79Epoch 35/50
8093/93 [==============================] - 2s 16ms/step - loss: 21.9862 - val_loss: 20.0650
81Epoch 36/50
8293/93 [==============================] - 1s 16ms/step - loss: 18.6333 - val_loss: 29.3458
83Epoch 37/50
8493/93 [==============================] - 1s 15ms/step - loss: 19.2257 - val_loss: 20.0688
85Epoch 38/50
8693/93 [==============================] - 1s 15ms/step - loss: 17.9420 - val_loss: 16.9885
87Epoch 39/50
8893/93 [==============================] - 2s 16ms/step - loss: 26.4506 - val_loss: 20.5957
89Epoch 40/50
9093/93 [==============================] - 1s 14ms/step - loss: 22.3560 - val_loss: 22.3593
91Epoch 41/50
9293/93 [==============================] - 1s 10ms/step - loss: 20.3805 - val_loss: 21.1551
93Epoch 42/50
9493/93 [==============================] - 1s 12ms/step - loss: 19.3720 - val_loss: 17.9814
95Epoch 43/50
9693/93 [==============================] - 1s 13ms/step - loss: 17.3375 - val_loss: 17.2637
97Epoch 44/50
9893/93 [==============================] - 2s 18ms/step - loss: 17.0008 - val_loss: 21.6005
99Epoch 45/50
10093/93 [==============================] - 1s 16ms/step - loss: 18.5636 - val_loss: 17.5460
101Epoch 46/50
10293/93 [==============================] - 1s 16ms/step - loss: 24.7443 - val_loss: 22.2168
103Epoch 47/50
10493/93 [==============================] - 1s 10ms/step - loss: 18.9257 - val_loss: 19.2673
105Epoch 48/50
10693/93 [==============================] - 1s 10ms/step - loss: 17.9717 - val_loss: 18.0424
107Epoch 49/50
10893/93 [==============================] - 1s 15ms/step - loss: 17.1485 - val_loss: 18.2311
109Epoch 50/50
11093/93 [==============================] - 1s 16ms/step - loss: 16.8210 - val_loss: 17.4355
check how validation behaves over time (epochs)
1import matplotlib.pyplot as plt
2plt.style.use("ggplot")
3plt.plot(np.arange(1, epochs + 1), H.history["loss"], label="Loss")
4plt.plot(np.arange(1, epochs + 1), H.history["val_loss"], label="val_loss")
5plt.xlabel("Epoch #")
6plt.ylabel("Loss")
7plt.legend()
8plt.show()
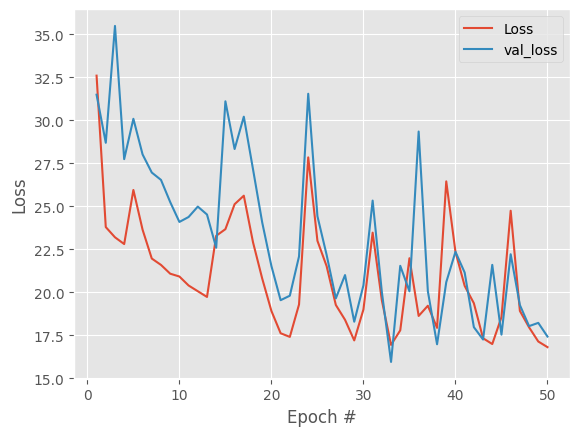
We see both training and validation loss are extremely noisy, with no clear improvement trend, and after ~15-20 epochs, improvements are just random fluctuations, not learning.
In these cases, it is useful to add regularization and decrease learning rate.
Regularization refers to a set of techniques used to reduce overfitting by discouraging overly complex models, typically by adding a penalty to the training objective that constrains model parameters. This encourages the model to learn more generalizable patterns rather than memorizing noise in the data.
L2 regularization (also known as weight decay) adds a penalty proportional to the sum of the squared model weights to the loss function, pushing weights toward smaller values without forcing them to zero. This helps stabilize training and promotes smoother, less sensitive models.
1from tensorflow.keras import regularizers
2
3def create_l2_autoencoder(neurons, input_shape, l2=1e-5):
4 input_layer = Input(shape=(input_shape,))
5
6 # Encoder layers
7 encoder_output = input_layer
8 for i in range(len(neurons)-1, 0, -1):
9 encoder_output = Dense(neurons[i], activation='elu',
10 kernel_regularizer=regularizers.l2(l2))(encoder_output)
11
12 # Hidden layer with a name
13 hidden_layer = Dense(neurons[0], activation='linear', name='hidden_layer',
14 kernel_regularizer=regularizers.l2(l2))(encoder_output)
15
16 # Decoder layers
17 decoder_output = hidden_layer
18 for i in range(1, len(neurons)):
19 decoder_output = Dense(neurons[i], activation='elu',
20 kernel_regularizer=regularizers.l2(l2))(decoder_output)
21
22 # Output layer
23 output_layer = Dense(input_shape, activation='linear')(decoder_output)
24
25 autoencoder = Model(inputs=input_layer, outputs=output_layer)
26 return autoencoder
re-initialize the model before running .fit() again to avoid keep training the same model
1autoencoder = create_l2_autoencoder(
2 neurons,
3 train_data.shape[1],
4 l2=1e-5
5)
6
7optimizer = tf.keras.optimizers.Adam(learning_rate=1e-3)
8autoencoder.compile(
9 optimizer=optimizer,
10 loss='mean_squared_error'
11)
12
13H = autoencoder.fit(
14 train_data,
15 train_data,
16 validation_data=(test_data, test_data),
17 batch_size=32,
18 epochs=epochs,
19 verbose=1
20)
1Epoch 1/50
293/93 [==============================] - 5s 16ms/step - loss: 26.6187 - val_loss: 27.4128
3Epoch 2/50
493/93 [==============================] - 1s 16ms/step - loss: 20.0764 - val_loss: 22.8442
5Epoch 3/50
693/93 [==============================] - 2s 17ms/step - loss: 16.3306 - val_loss: 19.9515
7Epoch 4/50
893/93 [==============================] - 1s 14ms/step - loss: 13.2374 - val_loss: 17.0505
9Epoch 5/50
1093/93 [==============================] - 2s 17ms/step - loss: 11.6092 - val_loss: 10.3541
11Epoch 6/50
1293/93 [==============================] - 2s 18ms/step - loss: 12.8784 - val_loss: 18.2110
13Epoch 7/50
1493/93 [==============================] - 1s 16ms/step - loss: 11.9601 - val_loss: 11.6533
15Epoch 8/50
1693/93 [==============================] - 1s 15ms/step - loss: 10.6742 - val_loss: 10.2509
17Epoch 9/50
1893/93 [==============================] - 1s 13ms/step - loss: 11.4126 - val_loss: 19.5551
19Epoch 10/50
2093/93 [==============================] - 2s 16ms/step - loss: 11.7762 - val_loss: 12.6366
21Epoch 11/50
2293/93 [==============================] - 1s 12ms/step - loss: 10.6223 - val_loss: 11.3330
23Epoch 12/50
2493/93 [==============================] - 2s 17ms/step - loss: 9.4480 - val_loss: 8.9790
25Epoch 13/50
2693/93 [==============================] - 2s 18ms/step - loss: 9.4324 - val_loss: 11.6798
27Epoch 14/50
2893/93 [==============================] - 2s 17ms/step - loss: 9.1592 - val_loss: 9.1531
29Epoch 15/50
3093/93 [==============================] - 1s 14ms/step - loss: 8.1193 - val_loss: 7.4658
31Epoch 16/50
3293/93 [==============================] - 1s 12ms/step - loss: 8.5194 - val_loss: 9.5459
33Epoch 17/50
3493/93 [==============================] - 2s 19ms/step - loss: 8.9682 - val_loss: 10.1431
35Epoch 18/50
3693/93 [==============================] - 2s 18ms/step - loss: 8.8097 - val_loss: 9.3160
37Epoch 19/50
3893/93 [==============================] - 1s 12ms/step - loss: 9.3907 - val_loss: 9.4668
39Epoch 20/50
4093/93 [==============================] - 2s 18ms/step - loss: 9.1025 - val_loss: 8.1897
41Epoch 21/50
4293/93 [==============================] - 1s 13ms/step - loss: 8.5533 - val_loss: 8.7505
43Epoch 22/50
4493/93 [==============================] - 2s 19ms/step - loss: 7.8477 - val_loss: 9.3622
45Epoch 23/50
4693/93 [==============================] - 1s 14ms/step - loss: 7.1622 - val_loss: 7.6489
47Epoch 24/50
4893/93 [==============================] - 2s 17ms/step - loss: 8.5040 - val_loss: 7.6554
49Epoch 25/50
5093/93 [==============================] - 2s 16ms/step - loss: 7.7141 - val_loss: 6.8418
51Epoch 26/50
5293/93 [==============================] - 1s 9ms/step - loss: 7.2177 - val_loss: 9.9998
53Epoch 27/50
5493/93 [==============================] - 1s 14ms/step - loss: 9.1925 - val_loss: 10.6097
55Epoch 28/50
5693/93 [==============================] - 1s 11ms/step - loss: 9.3420 - val_loss: 7.8157
57Epoch 29/50
5893/93 [==============================] - 1s 12ms/step - loss: 7.7143 - val_loss: 6.8660
59Epoch 30/50
6093/93 [==============================] - 2s 17ms/step - loss: 8.0236 - val_loss: 7.9869
61Epoch 31/50
6293/93 [==============================] - 2s 17ms/step - loss: 7.6223 - val_loss: 7.6642
63Epoch 32/50
6493/93 [==============================] - 1s 7ms/step - loss: 7.7311 - val_loss: 8.0535
65Epoch 33/50
6693/93 [==============================] - 1s 6ms/step - loss: 7.9626 - val_loss: 12.3125
67Epoch 34/50
6893/93 [==============================] - 1s 10ms/step - loss: 9.5091 - val_loss: 8.1515
69Epoch 35/50
7093/93 [==============================] - 1s 11ms/step - loss: 12.8089 - val_loss: 8.4810
71Epoch 36/50
7293/93 [==============================] - 1s 15ms/step - loss: 10.1668 - val_loss: 11.3264
73Epoch 37/50
7493/93 [==============================] - 1s 7ms/step - loss: 11.6888 - val_loss: 21.4890
75Epoch 38/50
7693/93 [==============================] - 1s 9ms/step - loss: 12.3693 - val_loss: 11.9961
77Epoch 39/50
7893/93 [==============================] - 1s 11ms/step - loss: 10.7251 - val_loss: 10.1258
79Epoch 40/50
8093/93 [==============================] - 1s 9ms/step - loss: 8.8597 - val_loss: 7.9398
81Epoch 41/50
8293/93 [==============================] - 1s 12ms/step - loss: 8.0705 - val_loss: 7.6555
83Epoch 42/50
8493/93 [==============================] - 1s 13ms/step - loss: 7.3514 - val_loss: 7.2101
85Epoch 43/50
8693/93 [==============================] - 1s 16ms/step - loss: 6.8367 - val_loss: 7.4991
87Epoch 44/50
8893/93 [==============================] - 2s 17ms/step - loss: 7.1375 - val_loss: 7.3583
89Epoch 45/50
9093/93 [==============================] - 1s 12ms/step - loss: 7.2660 - val_loss: 8.2415
91Epoch 46/50
9293/93 [==============================] - 2s 18ms/step - loss: 7.7464 - val_loss: 7.1394
93Epoch 47/50
9493/93 [==============================] - 1s 15ms/step - loss: 7.0515 - val_loss: 6.6802
95Epoch 48/50
9693/93 [==============================] - 2s 16ms/step - loss: 6.8640 - val_loss: 7.2763
97Epoch 49/50
9893/93 [==============================] - 1s 13ms/step - loss: 6.8215 - val_loss: 6.4784
99Epoch 50/50
10093/93 [==============================] - 1s 15ms/step - loss: 7.0084 - val_loss: 7.1828
and plot the losses
1plt.style.use("ggplot")
2N = len(H.history["loss"])
3plt.plot(np.arange(1, N + 1), H.history["loss"], label="Loss")
4plt.plot(np.arange(1, N + 1), H.history["val_loss"], label="val_loss")
5plt.xlabel("Epoch #")
6plt.ylabel("Loss")
7plt.legend()
8plt.show()
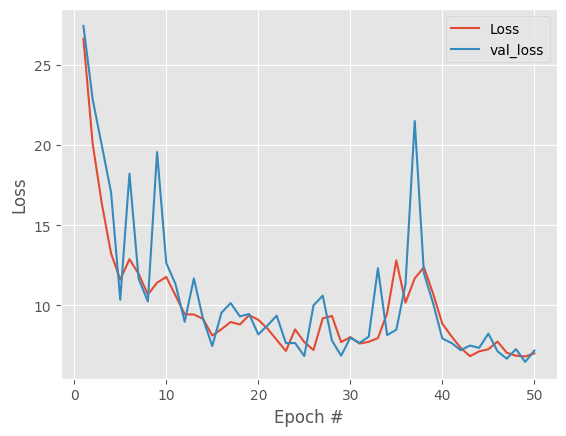
there is a clear improvement but we still have some large spikes and improvements after ~20 epochs are basically noise. With our data, one possible cause is data noise / dropout so we will input noise and repeat the whole process.
1from tensorflow.keras.layers import GaussianNoise
2
3def create_l2_noise_autoencoder(neurons, input_shape, l2=1e-5, noise_std=0.1):
4 input_layer = Input(shape=(input_shape,))
5 x = GaussianNoise(noise_std)(input_layer)
6
7 # Encoder layers
8 encoder_output = x
9 for i in range(len(neurons)-1, 0, -1):
10 encoder_output = Dense(neurons[i], activation='elu',
11 kernel_regularizer=regularizers.l2(l2))(encoder_output)
12
13 # Hidden layer with a name
14 hidden_layer = Dense(neurons[0], activation='linear', name='hidden_layer',
15 kernel_regularizer=regularizers.l2(l2))(encoder_output)
16
17 # Decoder layers
18 decoder_output = hidden_layer
19 for i in range(1, len(neurons)):
20 decoder_output = Dense(neurons[i], activation='elu',
21 kernel_regularizer=regularizers.l2(l2))(decoder_output)
22
23 # Output layer
24 output_layer = Dense(input_shape, activation='linear')(decoder_output)
25
26 autoencoder = Model(inputs=input_layer, outputs=output_layer)
27 return autoencoder
1autoencoder = create_l2_noise_autoencoder(
2 neurons,
3 train_data.shape[1],
4 l2=1e-5,
5 noise_std=0.1
6)
7
8optimizer = tf.keras.optimizers.Adam(learning_rate=1e-3)
9autoencoder.compile(
10 optimizer=optimizer,
11 loss='mean_squared_error'
12)
13
14H = autoencoder.fit(
15 train_data,
16 train_data,
17 validation_data=(test_data, test_data),
18 batch_size=32,
19 epochs=epochs,
20 verbose=1
21)
1Epoch 1/50
293/93 [==============================] - 4s 11ms/step - loss: 31.7372 - val_loss: 30.9056
3Epoch 2/50
493/93 [==============================] - 1s 16ms/step - loss: 24.1195 - val_loss: 30.1557
5Epoch 3/50
693/93 [==============================] - 1s 12ms/step - loss: 22.7737 - val_loss: 28.8883
7Epoch 4/50
893/93 [==============================] - 1s 12ms/step - loss: 22.4682 - val_loss: 28.5310
9Epoch 5/50
1093/93 [==============================] - 1s 13ms/step - loss: 22.3388 - val_loss: 27.8151
11Epoch 6/50
1293/93 [==============================] - 1s 16ms/step - loss: 22.3382 - val_loss: 31.0888
13Epoch 7/50
1493/93 [==============================] - 1s 15ms/step - loss: 22.1720 - val_loss: 27.5932
15Epoch 8/50
1693/93 [==============================] - 1s 13ms/step - loss: 21.6034 - val_loss: 27.3644
17Epoch 9/50
1893/93 [==============================] - 1s 9ms/step - loss: 22.4259 - val_loss: 31.4528
19Epoch 10/50
2093/93 [==============================] - 1s 15ms/step - loss: 22.8885 - val_loss: 28.0888
21Epoch 11/50
2293/93 [==============================] - 1s 12ms/step - loss: 21.8866 - val_loss: 26.8629
23Epoch 12/50
2493/93 [==============================] - 1s 15ms/step - loss: 21.5496 - val_loss: 27.7277
25Epoch 13/50
2693/93 [==============================] - 1s 14ms/step - loss: 21.3155 - val_loss: 26.2743
27Epoch 14/50
2893/93 [==============================] - 1s 13ms/step - loss: 22.5453 - val_loss: 25.7421
29Epoch 15/50
3093/93 [==============================] - 2s 17ms/step - loss: 20.7408 - val_loss: 25.2304
31Epoch 16/50
3293/93 [==============================] - 1s 12ms/step - loss: 20.3711 - val_loss: 25.2569
33Epoch 17/50
3493/93 [==============================] - 1s 16ms/step - loss: 19.8909 - val_loss: 23.9351
35Epoch 18/50
3693/93 [==============================] - 1s 14ms/step - loss: 19.7199 - val_loss: 23.9300
37Epoch 19/50
3893/93 [==============================] - 1s 14ms/step - loss: 19.2602 - val_loss: 23.2457
39Epoch 20/50
4093/93 [==============================] - 1s 15ms/step - loss: 18.9191 - val_loss: 22.8827
41Epoch 21/50
4293/93 [==============================] - 1s 14ms/step - loss: 18.5888 - val_loss: 23.1114
43Epoch 22/50
4493/93 [==============================] - 1s 16ms/step - loss: 18.4964 - val_loss: 22.9688
45Epoch 23/50
4693/93 [==============================] - 1s 11ms/step - loss: 18.6124 - val_loss: 22.2674
47Epoch 24/50
4893/93 [==============================] - 2s 17ms/step - loss: 17.7730 - val_loss: 23.0081
49Epoch 25/50
5093/93 [==============================] - 1s 9ms/step - loss: 18.0176 - val_loss: 21.7493
51Epoch 26/50
5293/93 [==============================] - 1s 9ms/step - loss: 17.4766 - val_loss: 20.5798
53Epoch 27/50
5493/93 [==============================] - 1s 15ms/step - loss: 17.2889 - val_loss: 20.2625
55Epoch 28/50
5693/93 [==============================] - 1s 16ms/step - loss: 17.4654 - val_loss: 23.4170
57Epoch 29/50
5893/93 [==============================] - 1s 10ms/step - loss: 16.9935 - val_loss: 20.6892
59Epoch 30/50
6093/93 [==============================] - 1s 13ms/step - loss: 28.9395 - val_loss: 68.7133
61Epoch 31/50
6293/93 [==============================] - 1s 15ms/step - loss: 27.4823 - val_loss: 27.9025
63Epoch 32/50
6493/93 [==============================] - 1s 14ms/step - loss: 21.1761 - val_loss: 24.7234
65Epoch 33/50
6693/93 [==============================] - 2s 16ms/step - loss: 19.8319 - val_loss: 24.1620
67Epoch 34/50
6893/93 [==============================] - 1s 14ms/step - loss: 19.3813 - val_loss: 23.4190
69Epoch 35/50
7093/93 [==============================] - 1s 14ms/step - loss: 18.8075 - val_loss: 22.6499
71Epoch 36/50
7293/93 [==============================] - 1s 12ms/step - loss: 19.0569 - val_loss: 22.2925
73Epoch 37/50
7493/93 [==============================] - 1s 10ms/step - loss: 18.8629 - val_loss: 22.6711
75Epoch 38/50
7693/93 [==============================] - 1s 15ms/step - loss: 16.1751 - val_loss: 16.3366
77Epoch 39/50
7893/93 [==============================] - 2s 18ms/step - loss: 14.4210 - val_loss: 15.0888
79Epoch 40/50
8093/93 [==============================] - 2s 16ms/step - loss: 13.0786 - val_loss: 13.6408
81Epoch 41/50
8293/93 [==============================] - 2s 17ms/step - loss: 12.8257 - val_loss: 14.2111
83Epoch 42/50
8493/93 [==============================] - 1s 15ms/step - loss: 12.2966 - val_loss: 13.4075
85Epoch 43/50
8693/93 [==============================] - 1s 11ms/step - loss: 11.4765 - val_loss: 11.9199
87Epoch 44/50
8893/93 [==============================] - 1s 14ms/step - loss: 12.7848 - val_loss: 14.4697
89Epoch 45/50
9093/93 [==============================] - 1s 10ms/step - loss: 18.1168 - val_loss: 17.5223
91Epoch 46/50
9293/93 [==============================] - 1s 14ms/step - loss: 14.1584 - val_loss: 15.3487
93Epoch 47/50
9493/93 [==============================] - 1s 14ms/step - loss: 12.4028 - val_loss: 13.6837
95Epoch 48/50
9693/93 [==============================] - 1s 16ms/step - loss: 11.8782 - val_loss: 12.1098
97Epoch 49/50
9893/93 [==============================] - 2s 16ms/step - loss: 10.6474 - val_loss: 11.5075
99Epoch 50/50
10093/93 [==============================] - 1s 11ms/step - loss: 10.1763 - val_loss: 11.3282
1plt.style.use("ggplot")
2N = len(H.history["loss"])
3plt.plot(np.arange(1, N + 1), H.history["loss"], label="Loss")
4plt.plot(np.arange(1, N + 1), H.history["val_loss"], label="val_loss")
5plt.xlabel("Epoch #")
6plt.ylabel("Loss")
7plt.legend()
8plt.show()
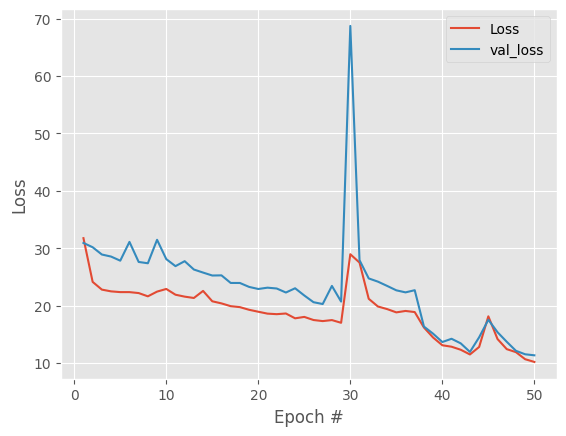
Good result, overall downward trend in both losses and track each other. Model is not overfitting. To remove the spikes in losses and get more stable embeddings lets try decreasing the learning rate.
1autoencoder = create_l2_noise_autoencoder(
2 neurons,
3 train_data.shape[1],
4 l2=1e-5,
5 noise_std=0.1
6)
7
8optimizer = tf.keras.optimizers.Adam(learning_rate=3e-4)
9autoencoder.compile(
10 optimizer=optimizer,
11 loss='mean_squared_error'
12)
13
14H = autoencoder.fit(
15 train_data,
16 train_data,
17 validation_data=(test_data, test_data),
18 batch_size=32,
19 epochs=epochs,
20 verbose=1
21)
1Epoch 1/50
293/93 [==============================] - 5s 15ms/step - loss: 38.6527 - val_loss: 39.9125
3Epoch 2/50
493/93 [==============================] - 1s 13ms/step - loss: 30.2958 - val_loss: 33.0952
5Epoch 3/50
693/93 [==============================] - 2s 18ms/step - loss: 24.9085 - val_loss: 32.5780
7Epoch 4/50
893/93 [==============================] - 1s 13ms/step - loss: 23.2918 - val_loss: 29.3479
9Epoch 5/50
1093/93 [==============================] - 1s 15ms/step - loss: 22.1767 - val_loss: 28.1120
11Epoch 6/50
1293/93 [==============================] - 1s 16ms/step - loss: 21.7849 - val_loss: 27.8594
13Epoch 7/50
1493/93 [==============================] - 1s 12ms/step - loss: 21.6366 - val_loss: 27.7357
15Epoch 8/50
1693/93 [==============================] - 1s 15ms/step - loss: 21.4320 - val_loss: 27.2052
17Epoch 9/50
1893/93 [==============================] - 1s 14ms/step - loss: 21.4469 - val_loss: 26.7283
19Epoch 10/50
2093/93 [==============================] - 2s 16ms/step - loss: 20.8234 - val_loss: 26.0348
21Epoch 11/50
2293/93 [==============================] - 2s 17ms/step - loss: 20.5192 - val_loss: 25.8189
23Epoch 12/50
2493/93 [==============================] - 1s 12ms/step - loss: 20.2020 - val_loss: 24.7882
25Epoch 13/50
2693/93 [==============================] - 1s 12ms/step - loss: 20.1402 - val_loss: 24.2947
27Epoch 14/50
2893/93 [==============================] - 1s 14ms/step - loss: 19.4716 - val_loss: 24.3214
29Epoch 15/50
3093/93 [==============================] - 2s 18ms/step - loss: 19.7134 - val_loss: 25.5420
31Epoch 16/50
3293/93 [==============================] - 1s 16ms/step - loss: 19.3621 - val_loss: 19.9822
33Epoch 17/50
3493/93 [==============================] - 1s 13ms/step - loss: 17.4665 - val_loss: 22.8801
35Epoch 18/50
3693/93 [==============================] - 1s 13ms/step - loss: 18.6423 - val_loss: 22.3084
37Epoch 19/50
3893/93 [==============================] - 1s 15ms/step - loss: 17.2165 - val_loss: 22.0067
39Epoch 20/50
4093/93 [==============================] - 1s 10ms/step - loss: 16.5752 - val_loss: 21.4383
41Epoch 21/50
4293/93 [==============================] - 1s 12ms/step - loss: 16.1724 - val_loss: 20.6535
43Epoch 22/50
4493/93 [==============================] - 1s 12ms/step - loss: 15.7307 - val_loss: 20.1583
45Epoch 23/50
4693/93 [==============================] - 1s 15ms/step - loss: 15.4491 - val_loss: 20.5153
47Epoch 24/50
4893/93 [==============================] - 1s 8ms/step - loss: 15.2299 - val_loss: 19.4298
49Epoch 25/50
5093/93 [==============================] - 1s 14ms/step - loss: 14.6663 - val_loss: 19.3464
51Epoch 26/50
5293/93 [==============================] - 1s 12ms/step - loss: 14.2106 - val_loss: 18.0098
53Epoch 27/50
5493/93 [==============================] - 1s 15ms/step - loss: 13.7767 - val_loss: 18.0012
55Epoch 28/50
5693/93 [==============================] - 1s 13ms/step - loss: 14.1194 - val_loss: 16.8258
57Epoch 29/50
5893/93 [==============================] - 1s 15ms/step - loss: 14.2060 - val_loss: 17.3795
59Epoch 30/50
6093/93 [==============================] - 1s 14ms/step - loss: 13.5694 - val_loss: 17.7462
61Epoch 31/50
6293/93 [==============================] - 2s 16ms/step - loss: 13.1289 - val_loss: 16.7373
63Epoch 32/50
6493/93 [==============================] - 1s 9ms/step - loss: 12.7070 - val_loss: 17.3542
65Epoch 33/50
6693/93 [==============================] - 1s 15ms/step - loss: 14.6508 - val_loss: 16.4855
67Epoch 34/50
6893/93 [==============================] - 1s 13ms/step - loss: 12.7817 - val_loss: 15.9853
69Epoch 35/50
7093/93 [==============================] - 1s 16ms/step - loss: 12.8153 - val_loss: 15.8276
71Epoch 36/50
7293/93 [==============================] - 1s 15ms/step - loss: 13.0446 - val_loss: 16.2531
73Epoch 37/50
7493/93 [==============================] - 1s 16ms/step - loss: 14.1267 - val_loss: 18.5714
75Epoch 38/50
7693/93 [==============================] - 1s 14ms/step - loss: 14.6227 - val_loss: 21.0582
77Epoch 39/50
7893/93 [==============================] - 1s 14ms/step - loss: 15.4332 - val_loss: 17.4142
79Epoch 40/50
8093/93 [==============================] - 1s 12ms/step - loss: 15.7435 - val_loss: 19.7408
81Epoch 41/50
8293/93 [==============================] - 1s 15ms/step - loss: 13.3249 - val_loss: 24.3964
83Epoch 42/50
8493/93 [==============================] - 1s 12ms/step - loss: 14.4557 - val_loss: 14.6062
85Epoch 43/50
8693/93 [==============================] - 1s 14ms/step - loss: 11.7363 - val_loss: 13.9266
87Epoch 44/50
8893/93 [==============================] - 1s 13ms/step - loss: 11.4413 - val_loss: 16.5276
89Epoch 45/50
9093/93 [==============================] - 1s 13ms/step - loss: 11.2720 - val_loss: 15.1880
91Epoch 46/50
9293/93 [==============================] - 1s 14ms/step - loss: 10.0483 - val_loss: 12.4647
93Epoch 47/50
9493/93 [==============================] - 1s 16ms/step - loss: 11.4013 - val_loss: 14.8307
95Epoch 48/50
9693/93 [==============================] - 1s 11ms/step - loss: 11.3730 - val_loss: 13.1132
97Epoch 49/50
9893/93 [==============================] - 1s 12ms/step - loss: 10.5250 - val_loss: 11.7176
99Epoch 50/50
10093/93 [==============================] - 1s 15ms/step - loss: 9.7135 - val_loss: 14.0300
1plt.style.use("ggplot")
2N = len(H.history["loss"])
3plt.plot(np.arange(1, N + 1), H.history["loss"], label="Loss")
4plt.plot(np.arange(1, N + 1), H.history["val_loss"], label="val_loss")
5plt.xlabel("Epoch #")
6plt.ylabel("Loss")
7plt.legend()
8plt.show()
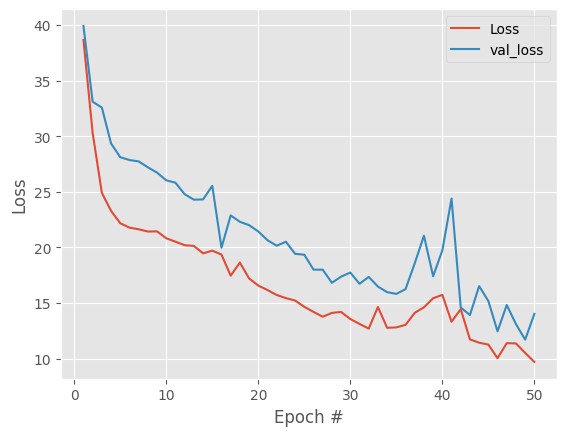
at this point we can extract the hidden layer or bottleneck to use it as dimensional reduction coordinates in our pancreatic cells sce object.
1hidden_layer_model = Model(inputs=autoencoder.input, outputs=autoencoder.get_layer('hidden_layer').output)
2hidden_layer_values = hidden_layer_model.predict(input_data)
1116/116 [==============================] - 0s 1ms/step
1print("Shape of hidden layer values:", hidden_layer_values.shape)
1Shape of hidden layer values: (3696, 2)
and save it to disk to open it outside docker
1hidden_layer_array = np.array(hidden_layer_values)
2hidden_layer_df = pd.DataFrame(hidden_layer_array, columns=[f'Autoencoder_{i}' for i in range(hidden_layer_array.shape[1])])
3csv_output_path = 'hidden_layer.docker.1024.4.hidden2.earlystop.fixPreprocessing.csv'
4hidden_layer_df.to_csv(csv_output_path, index=False)
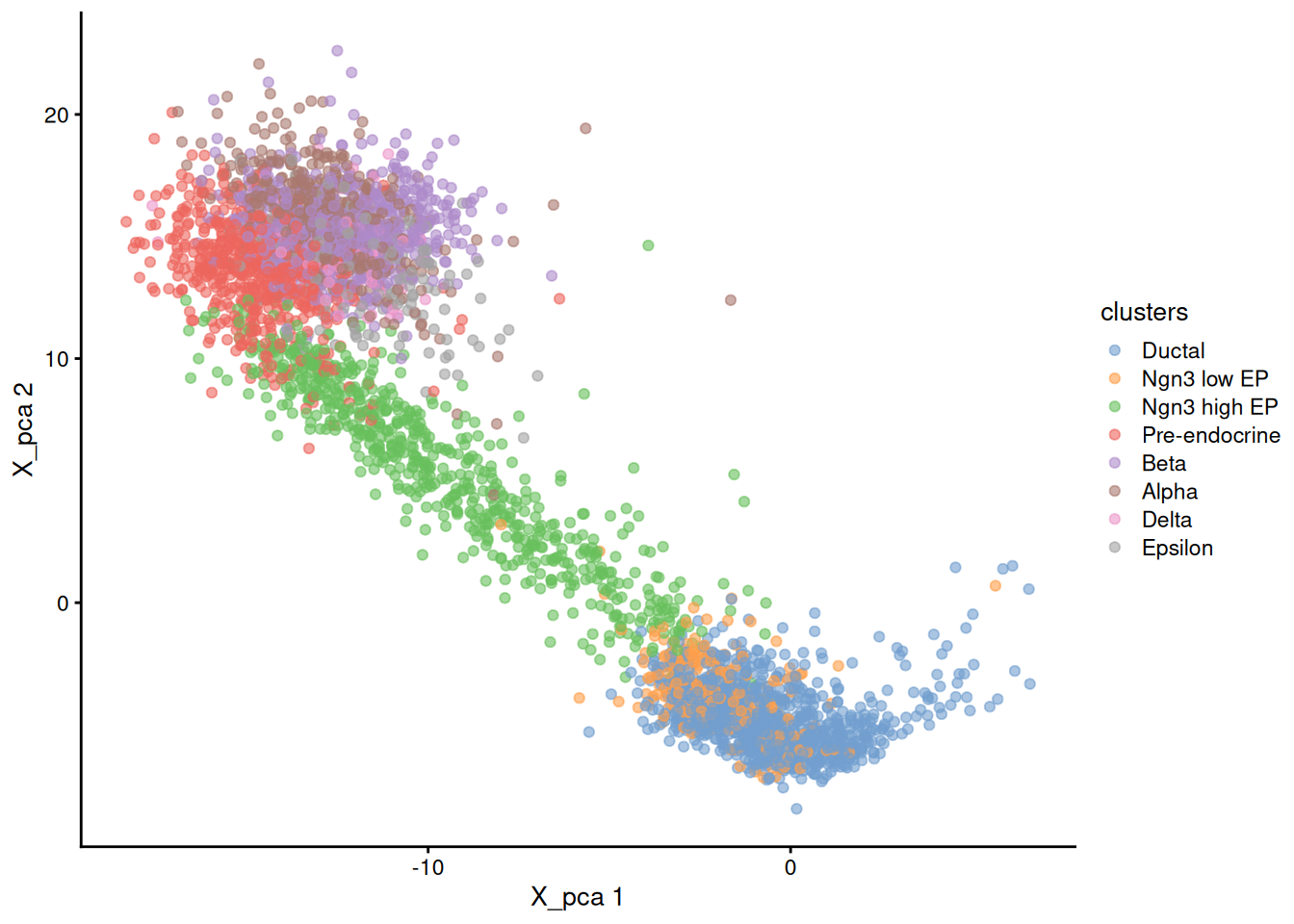
now let's get back to R (or python) outside docker and plot the original PCA
1library(scater)
2plotReducedDim( sce, dimred = 'X_pca', colour_by='clusters' )
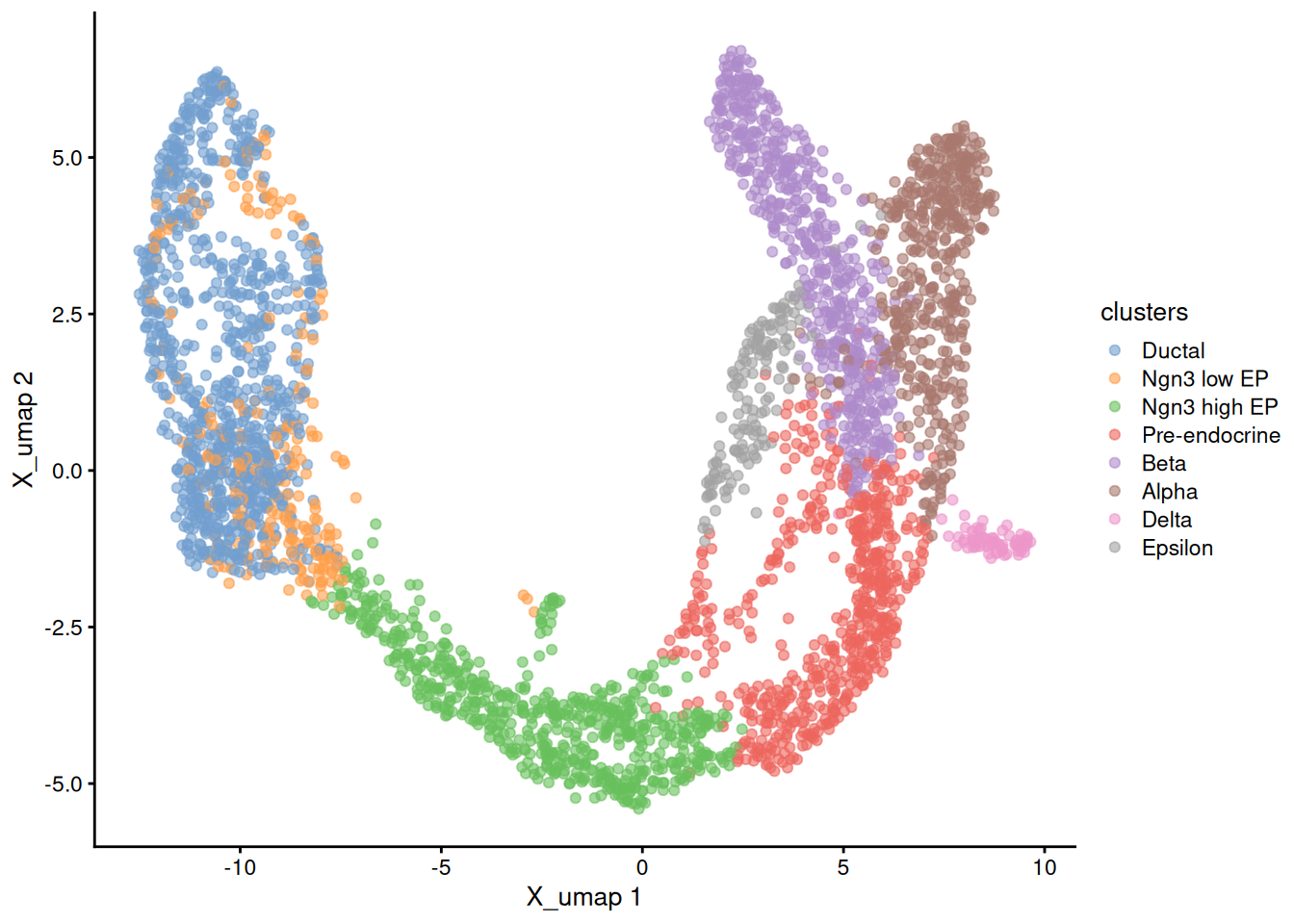
and the original UMAP
1plotReducedDim( sce, dimred = 'X_umap', colour_by='clusters' )
now we read the autoencoder bottleneck data and add it to the SCE reducedDim slot.
1autoencoder.df <- read.csv( 'hidden_layer.docker.1024.4.hidden2.earlystop.fixPreprocessing.csv' )
2rownames(autoencoder.df) <- colnames(sce)
3reducedDim( sce, 'AE' ) <- autoencoder.df
now plot the results
1plotReducedDim( sce, dimred = 'AE', colour_by='clusters' )
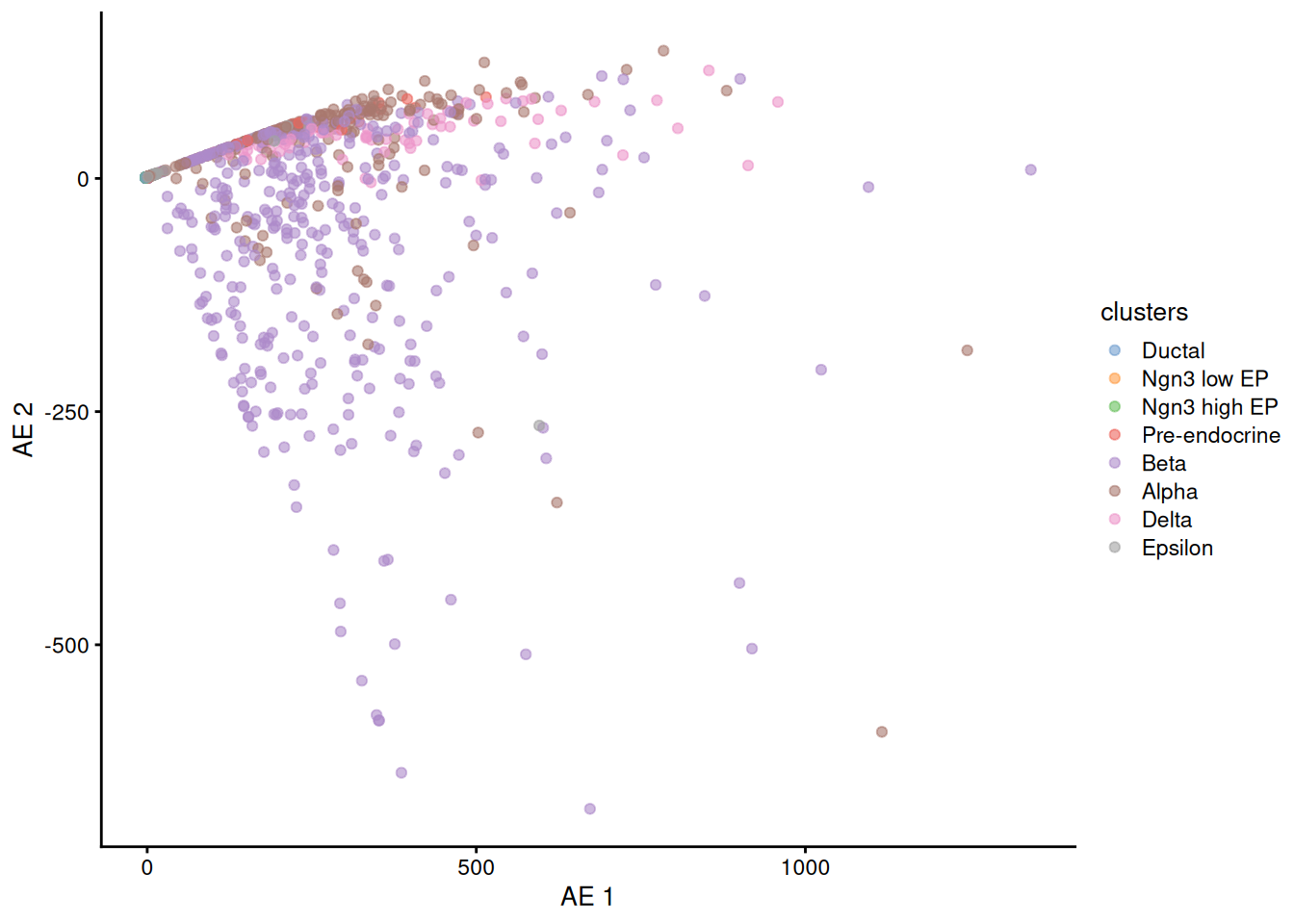
It seems our basic AE is not good enough to be used directly to plot the cells.
Next we will try to use the same autoencoder approach but defining a 50 neuron bottleneck that we will use later as the basis to the tSNE and UMAP representations as we normally do with PCA.
Open a new jupyter notebook inside the docker and do the same until defining the number of neurons per layer
1neurons = [50] + [2**x for x in range(6,11)]
2neurons
1[50, 64, 128, 256, 512, 1024]
learning rate and the number of epochs
1custom_learning_rate = 0.001
2epochs = 50
autoencoder function
1def create_autoencoder(neurons, input_shape):
2 input_layer = Input(shape=(input_shape,))
3
4 # Encoder layers
5 encoder_output = input_layer
6 for i in range(len(neurons)-1, 0, -1):
7 encoder_output = Dense(neurons[i], activation='elu')(encoder_output)
8
9 # Hidden layer with a name
10 hidden_layer = Dense(neurons[0], activation='linear', name='hidden_layer')(encoder_output)
11
12 # Decoder layers
13 decoder_output = hidden_layer
14 for i in range(1, len(neurons)):
15 decoder_output = Dense(neurons[i], activation='elu')(decoder_output)
16
17 # Output layer
18 output_layer = Dense(input_shape, activation='linear')(decoder_output)
19 # output_layer = Dense(input_shape, activation='sigmoid')(decoder_output) # changed because input goes from -1 to 1
20
21 autoencoder = Model(inputs=input_layer, outputs=output_layer)
22 return autoencoder
autoencoder set up
1autoencoder = create_autoencoder(neurons, train_data.shape[1])
12025-12-17 17:28:28.376131: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
22025-12-17 17:28:28.437362: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
32025-12-17 17:28:28.440064: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
42025-12-17 17:28:28.443025: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
52025-12-17 17:28:28.445118: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
62025-12-17 17:28:28.450453: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
72025-12-17 17:28:28.598267: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
82025-12-17 17:28:28.599087: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
92025-12-17 17:28:28.599764: I external/local_xla/xla/stream_executor/cuda/cuda_executor.cc:901] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
102025-12-17 17:28:28.600633: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1929] Created device /job:localhost/replica:0/task:0/device:GPU:0 with 6002 MB memory: -> device: 0, name: NVIDIA GeForce RTX 2070 SUPER, pci bus id: 0000:01:00.0, compute capability: 7.5
optimizer and compilation
1optimizer = tf.keras.optimizers.Adam(learning_rate=custom_learning_rate)
2autoencoder.compile(optimizer=optimizer, loss='mean_squared_error')
model summary
1autoencoder.summary()
1Model: "model"
2_________________________________________________________________
3 Layer (type) Output Shape Param #
4=================================================================
5 input_1 (InputLayer) [(None, 2000)] 0
6
7 dense (Dense) (None, 1024) 2049024
8
9 dense_1 (Dense) (None, 512) 524800
10
11 dense_2 (Dense) (None, 256) 131328
12
13 dense_3 (Dense) (None, 128) 32896
14
15 dense_4 (Dense) (None, 64) 8256
16
17 hidden_layer (Dense) (None, 50) 3250
18
19 dense_5 (Dense) (None, 64) 3264
20
21 dense_6 (Dense) (None, 128) 8320
22
23 dense_7 (Dense) (None, 256) 33024
24
25 dense_8 (Dense) (None, 512) 131584
26
27 dense_9 (Dense) (None, 1024) 525312
28
29 dense_10 (Dense) (None, 2000) 2050000
30
31=================================================================
32Total params: 5501058 (20.98 MB)
33Trainable params: 5501058 (20.98 MB)
34Non-trainable params: 0 (0.00 Byte)
35_________________________________________________________________
training
1H = autoencoder.fit(x = train_data, y = train_data,
2 validation_data=(test_data, test_data),
3 batch_size = 32, epochs = epochs, verbose = 1)
1Epoch 1/50
2
3
42025-12-17 17:29:12.731974: I external/local_xla/xla/service/service.cc:168] XLA service 0x76a550615c30 initialized for platform CUDA (this does not guarantee that XLA will be used). Devices:
52025-12-17 17:29:12.731994: I external/local_xla/xla/service/service.cc:176] StreamExecutor device (0): NVIDIA GeForce RTX 2070 SUPER, Compute Capability 7.5
62025-12-17 17:29:12.741006: I tensorflow/compiler/mlir/tensorflow/utils/dump_mlir_util.cc:269] disabling MLIR crash reproducer, set env var `MLIR_CRASH_REPRODUCER_DIRECTORY` to enable.
72025-12-17 17:29:12.761897: I external/local_xla/xla/stream_executor/cuda/cuda_dnn.cc:454] Loaded cuDNN version 8906
8WARNING: All log messages before absl::InitializeLog() is called are written to STDERR
9I0000 00:00:1765992552.816020 271 device_compiler.h:186] Compiled cluster using XLA! This line is logged at most once for the lifetime of the process.
10
11
1293/93 [==============================] - 4s 11ms/step - loss: 20.1129 - val_loss: 10.9373
13Epoch 2/50
1493/93 [==============================] - 1s 13ms/step - loss: 13.1907 - val_loss: 11.7414
15Epoch 3/50
1693/93 [==============================] - 1s 14ms/step - loss: 9.1359 - val_loss: 12.6089
17Epoch 4/50
1893/93 [==============================] - 1s 12ms/step - loss: 10.0069 - val_loss: 7.0463
19Epoch 5/50
2093/93 [==============================] - 1s 13ms/step - loss: 7.0336 - val_loss: 7.4595
21Epoch 6/50
2293/93 [==============================] - 1s 9ms/step - loss: 7.5198 - val_loss: 9.9332
23Epoch 7/50
2493/93 [==============================] - 1s 8ms/step - loss: 7.8503 - val_loss: 8.0087
25Epoch 8/50
2693/93 [==============================] - 1s 12ms/step - loss: 7.7250 - val_loss: 12.2940
27Epoch 9/50
2893/93 [==============================] - 1s 11ms/step - loss: 11.0793 - val_loss: 18.4790
29Epoch 10/50
3093/93 [==============================] - 1s 9ms/step - loss: 11.1037 - val_loss: 15.8162
31Epoch 11/50
3293/93 [==============================] - 1s 12ms/step - loss: 9.3877 - val_loss: 9.6597
33Epoch 12/50
3493/93 [==============================] - 1s 12ms/step - loss: 6.8708 - val_loss: 6.3998
35Epoch 13/50
3693/93 [==============================] - 1s 12ms/step - loss: 5.1222 - val_loss: 6.6202
37Epoch 14/50
3893/93 [==============================] - 1s 12ms/step - loss: 5.3738 - val_loss: 5.1922
39Epoch 15/50
4093/93 [==============================] - 1s 8ms/step - loss: 5.0246 - val_loss: 7.0998
41Epoch 16/50
4293/93 [==============================] - 1s 11ms/step - loss: 5.5951 - val_loss: 6.0730
43Epoch 17/50
4493/93 [==============================] - 1s 9ms/step - loss: 4.4057 - val_loss: 4.6819
45Epoch 18/50
4693/93 [==============================] - 1s 9ms/step - loss: 3.9670 - val_loss: 4.3718
47Epoch 19/50
4893/93 [==============================] - 1s 12ms/step - loss: 4.0684 - val_loss: 5.0374
49Epoch 20/50
5093/93 [==============================] - 1s 7ms/step - loss: 4.8767 - val_loss: 4.8057
51Epoch 21/50
5293/93 [==============================] - 1s 13ms/step - loss: 6.6046 - val_loss: 10.8622
53Epoch 22/50
5493/93 [==============================] - 1s 13ms/step - loss: 6.4166 - val_loss: 10.4268
55Epoch 23/50
5693/93 [==============================] - 1s 10ms/step - loss: 5.2859 - val_loss: 6.9333
57Epoch 24/50
5893/93 [==============================] - 1s 13ms/step - loss: 4.6556 - val_loss: 5.1214
59Epoch 25/50
6093/93 [==============================] - 1s 8ms/step - loss: 4.4241 - val_loss: 6.0715
61Epoch 26/50
6293/93 [==============================] - 1s 12ms/step - loss: 4.0185 - val_loss: 4.7227
63Epoch 27/50
6493/93 [==============================] - 1s 8ms/step - loss: 3.9552 - val_loss: 5.1046
65Epoch 28/50
6693/93 [==============================] - 1s 13ms/step - loss: 4.0014 - val_loss: 4.4248
67Epoch 29/50
6893/93 [==============================] - 1s 13ms/step - loss: 3.8944 - val_loss: 9.6719
69Epoch 30/50
7093/93 [==============================] - 1s 13ms/step - loss: 4.9995 - val_loss: 7.0005
71Epoch 31/50
7293/93 [==============================] - 1s 10ms/step - loss: 6.1998 - val_loss: 6.9595
73Epoch 32/50
7493/93 [==============================] - 1s 15ms/step - loss: 5.2774 - val_loss: 6.0492
75Epoch 33/50
7693/93 [==============================] - 1s 9ms/step - loss: 8.4152 - val_loss: 9.5338
77Epoch 34/50
7893/93 [==============================] - 1s 9ms/step - loss: 6.7191 - val_loss: 6.3981
79Epoch 35/50
8093/93 [==============================] - 1s 12ms/step - loss: 5.2410 - val_loss: 9.8374
81Epoch 36/50
8293/93 [==============================] - 1s 11ms/step - loss: 4.7739 - val_loss: 5.9090
83Epoch 37/50
8493/93 [==============================] - 1s 12ms/step - loss: 4.4791 - val_loss: 5.0151
85Epoch 38/50
8693/93 [==============================] - 1s 12ms/step - loss: 4.1940 - val_loss: 4.9561
87Epoch 39/50
8893/93 [==============================] - 1s 11ms/step - loss: 3.8426 - val_loss: 4.9457
89Epoch 40/50
9093/93 [==============================] - 1s 7ms/step - loss: 4.2509 - val_loss: 4.4218
91Epoch 41/50
9293/93 [==============================] - 1s 11ms/step - loss: 3.6131 - val_loss: 4.6667
93Epoch 42/50
9493/93 [==============================] - 1s 11ms/step - loss: 3.4824 - val_loss: 3.9728
95Epoch 43/50
9693/93 [==============================] - 1s 13ms/step - loss: 3.2685 - val_loss: 4.2212
97Epoch 44/50
9893/93 [==============================] - 1s 11ms/step - loss: 3.4520 - val_loss: 4.1716
99Epoch 45/50
10093/93 [==============================] - 1s 12ms/step - loss: 3.1752 - val_loss: 3.7729
101Epoch 46/50
10293/93 [==============================] - 1s 9ms/step - loss: 3.0366 - val_loss: 3.9370
103Epoch 47/50
10493/93 [==============================] - 1s 12ms/step - loss: 3.0311 - val_loss: 4.2225
105Epoch 48/50
10693/93 [==============================] - 1s 12ms/step - loss: 3.0429 - val_loss: 3.6647
107Epoch 49/50
10893/93 [==============================] - 1s 9ms/step - loss: 3.7111 - val_loss: 5.3528
109Epoch 50/50
11093/93 [==============================] - 1s 6ms/step - loss: 3.5047 - val_loss: 4.9664
plot
1import matplotlib.pyplot as plt
2plt.style.use("ggplot")
3plt.plot(np.arange(1, epochs + 1), H.history["loss"], label="Loss")
4plt.plot(np.arange(1, epochs + 1), H.history["val_loss"], label="val_loss")
5plt.xlabel("Epoch #")
6plt.ylabel("Loss")
7plt.legend()
8plt.show()
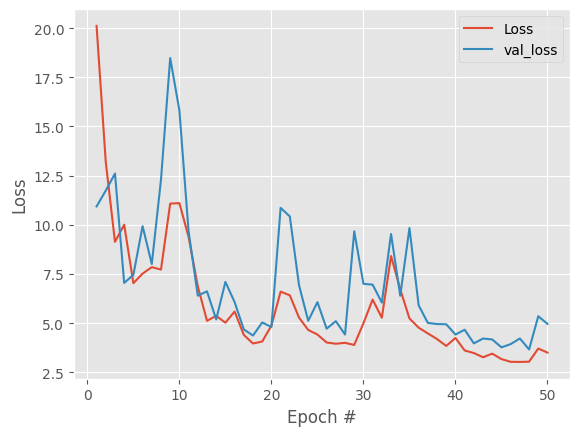
add regularization and decreate learning rate
1from tensorflow.keras import regularizers
2
3def create_l2_autoencoder(neurons, input_shape, l2=1e-5):
4 input_layer = Input(shape=(input_shape,))
5
6 # Encoder layers
7 encoder_output = input_layer
8 for i in range(len(neurons)-1, 0, -1):
9 encoder_output = Dense(neurons[i], activation='elu',
10 kernel_regularizer=regularizers.l2(l2))(encoder_output)
11
12 # Hidden layer with a name
13 hidden_layer = Dense(neurons[0], activation='linear', name='hidden_layer',
14 kernel_regularizer=regularizers.l2(l2))(encoder_output)
15
16 # Decoder layers
17 decoder_output = hidden_layer
18 for i in range(1, len(neurons)):
19 decoder_output = Dense(neurons[i], activation='elu',
20 kernel_regularizer=regularizers.l2(l2))(decoder_output)
21
22 # Output layer
23 output_layer = Dense(input_shape, activation='linear')(decoder_output)
24
25 autoencoder = Model(inputs=input_layer, outputs=output_layer)
26 return autoencoder
re-initialize the model
1autoencoder = create_l2_autoencoder(
2 neurons,
3 train_data.shape[1],
4 l2=1e-5
5)
6
7optimizer = tf.keras.optimizers.Adam(learning_rate=1e-3)
8autoencoder.compile(
9 optimizer=optimizer,
10 loss='mean_squared_error'
11)
12
13H = autoencoder.fit(
14 train_data,
15 train_data,
16 validation_data=(test_data, test_data),
17 batch_size=32,
18 epochs=epochs,
19 verbose=1
20)
1Epoch 1/50
293/93 [==============================] - 3s 13ms/step - loss: 19.0678 - val_loss: 18.4396
3Epoch 2/50
493/93 [==============================] - 1s 11ms/step - loss: 11.2227 - val_loss: 9.2450
5Epoch 3/50
693/93 [==============================] - 1s 11ms/step - loss: 8.4250 - val_loss: 7.0933
7Epoch 4/50
893/93 [==============================] - 1s 11ms/step - loss: 7.8562 - val_loss: 15.4034
9Epoch 5/50
1093/93 [==============================] - 1s 6ms/step - loss: 7.9933 - val_loss: 10.1543
11Epoch 6/50
1293/93 [==============================] - 1s 10ms/step - loss: 8.9621 - val_loss: 12.6527
13Epoch 7/50
1493/93 [==============================] - 1s 11ms/step - loss: 7.9506 - val_loss: 6.0607
15Epoch 8/50
1693/93 [==============================] - 1s 11ms/step - loss: 6.2425 - val_loss: 5.4726
17Epoch 9/50
1893/93 [==============================] - 1s 9ms/step - loss: 5.3090 - val_loss: 5.7768
19Epoch 10/50
2093/93 [==============================] - 1s 9ms/step - loss: 7.0954 - val_loss: 8.2602
21Epoch 11/50
2293/93 [==============================] - 1s 13ms/step - loss: 7.0199 - val_loss: 11.3992
23Epoch 12/50
2493/93 [==============================] - 1s 13ms/step - loss: 7.4499 - val_loss: 7.3681
25Epoch 13/50
2693/93 [==============================] - 1s 12ms/step - loss: 6.8692 - val_loss: 7.4825
27Epoch 14/50
2893/93 [==============================] - 1s 13ms/step - loss: 5.5452 - val_loss: 6.1778
29Epoch 15/50
3093/93 [==============================] - 1s 12ms/step - loss: 5.4449 - val_loss: 5.8070
31Epoch 16/50
3293/93 [==============================] - 1s 8ms/step - loss: 5.0635 - val_loss: 8.8165
33Epoch 17/50
3493/93 [==============================] - 1s 7ms/step - loss: 6.2696 - val_loss: 8.8737
35Epoch 18/50
3693/93 [==============================] - 1s 11ms/step - loss: 6.3253 - val_loss: 8.1490
37Epoch 19/50
3893/93 [==============================] - 1s 8ms/step - loss: 6.2105 - val_loss: 5.5200
39Epoch 20/50
4093/93 [==============================] - 1s 12ms/step - loss: 4.9530 - val_loss: 7.6524
41Epoch 21/50
4293/93 [==============================] - 1s 12ms/step - loss: 5.0426 - val_loss: 6.6516
43Epoch 22/50
4493/93 [==============================] - 1s 7ms/step - loss: 5.5303 - val_loss: 6.0581
45Epoch 23/50
4693/93 [==============================] - 1s 12ms/step - loss: 5.0887 - val_loss: 6.1520
47Epoch 24/50
4893/93 [==============================] - 1s 11ms/step - loss: 4.7274 - val_loss: 5.2073
49Epoch 25/50
5093/93 [==============================] - 1s 11ms/step - loss: 4.5191 - val_loss: 5.6213
51Epoch 26/50
5293/93 [==============================] - 1s 7ms/step - loss: 5.3404 - val_loss: 5.1017
53Epoch 27/50
5493/93 [==============================] - 1s 8ms/step - loss: 4.5175 - val_loss: 5.4251
55Epoch 28/50
5693/93 [==============================] - 1s 10ms/step - loss: 3.9859 - val_loss: 7.9162
57Epoch 29/50
5893/93 [==============================] - 1s 11ms/step - loss: 4.2546 - val_loss: 4.4417
59Epoch 30/50
6093/93 [==============================] - 1s 8ms/step - loss: 3.5685 - val_loss: 5.0945
61Epoch 31/50
6293/93 [==============================] - 1s 9ms/step - loss: 3.4755 - val_loss: 4.2805
63Epoch 32/50
6493/93 [==============================] - 1s 10ms/step - loss: 3.8756 - val_loss: 4.5240
65Epoch 33/50
6693/93 [==============================] - 1s 10ms/step - loss: 3.9401 - val_loss: 3.7048
67Epoch 34/50
6893/93 [==============================] - 1s 10ms/step - loss: 3.6122 - val_loss: 4.7491
69Epoch 35/50
7093/93 [==============================] - 1s 9ms/step - loss: 3.4352 - val_loss: 4.3423
71Epoch 36/50
7293/93 [==============================] - 1s 9ms/step - loss: 3.5778 - val_loss: 8.0054
73Epoch 37/50
7493/93 [==============================] - 1s 11ms/step - loss: 4.2836 - val_loss: 4.5677
75Epoch 38/50
7693/93 [==============================] - 1s 7ms/step - loss: 3.5487 - val_loss: 3.8854
77Epoch 39/50
7893/93 [==============================] - 1s 12ms/step - loss: 3.1797 - val_loss: 3.9341
79Epoch 40/50
8093/93 [==============================] - 1s 13ms/step - loss: 3.9631 - val_loss: 5.6835
81Epoch 41/50
8293/93 [==============================] - 1s 8ms/step - loss: 3.6220 - val_loss: 4.6448
83Epoch 42/50
8493/93 [==============================] - 1s 8ms/step - loss: 3.2747 - val_loss: 3.7277
85Epoch 43/50
8693/93 [==============================] - 1s 8ms/step - loss: 2.9811 - val_loss: 4.0839
87Epoch 44/50
8893/93 [==============================] - 1s 10ms/step - loss: 3.9734 - val_loss: 5.5020
89Epoch 45/50
9093/93 [==============================] - 1s 11ms/step - loss: 4.3734 - val_loss: 4.4314
91Epoch 46/50
9293/93 [==============================] - 1s 10ms/step - loss: 3.8605 - val_loss: 4.8687
93Epoch 47/50
9493/93 [==============================] - 1s 13ms/step - loss: 3.7208 - val_loss: 4.3981
95Epoch 48/50
9693/93 [==============================] - 1s 12ms/step - loss: 3.7345 - val_loss: 4.3941
97Epoch 49/50
9893/93 [==============================] - 1s 10ms/step - loss: 3.1808 - val_loss: 3.9066
99Epoch 50/50
10093/93 [==============================] - 1s 8ms/step - loss: 3.2613 - val_loss: 4.5659
1plt.style.use("ggplot")
2N = len(H.history["loss"])
3plt.plot(np.arange(1, N + 1), H.history["loss"], label="Loss")
4plt.plot(np.arange(1, N + 1), H.history["val_loss"], label="val_loss")
5plt.xlabel("Epoch #")
6plt.ylabel("Loss")
7plt.legend()
8plt.show()
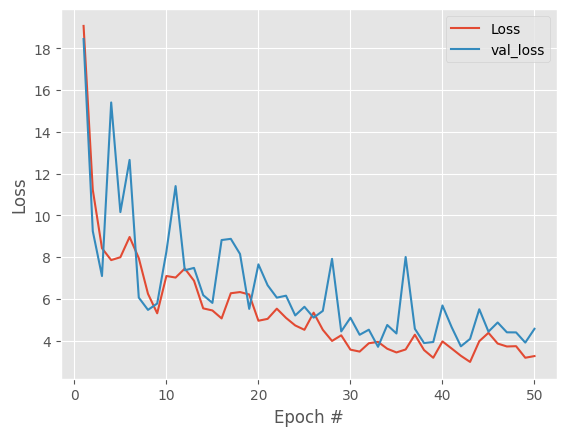
add noise
1from tensorflow.keras.layers import GaussianNoise
2
3def create_l2_noise_autoencoder(neurons, input_shape, l2=1e-5, noise_std=0.1):
4 input_layer = Input(shape=(input_shape,))
5 x = GaussianNoise(noise_std)(input_layer)
6
7 # Encoder layers
8 encoder_output = x
9 for i in range(len(neurons)-1, 0, -1):
10 encoder_output = Dense(neurons[i], activation='elu',
11 kernel_regularizer=regularizers.l2(l2))(encoder_output)
12
13 # Hidden layer with a name
14 hidden_layer = Dense(neurons[0], activation='linear', name='hidden_layer',
15 kernel_regularizer=regularizers.l2(l2))(encoder_output)
16
17 # Decoder layers
18 decoder_output = hidden_layer
19 for i in range(1, len(neurons)):
20 decoder_output = Dense(neurons[i], activation='elu',
21 kernel_regularizer=regularizers.l2(l2))(decoder_output)
22
23 # Output layer
24 output_layer = Dense(input_shape, activation='linear')(decoder_output)
25
26 autoencoder = Model(inputs=input_layer, outputs=output_layer)
27 return autoencoder
1autoencoder = create_l2_noise_autoencoder(
2 neurons,
3 train_data.shape[1],
4 l2=1e-5,
5 noise_std=0.1
6)
7
8optimizer = tf.keras.optimizers.Adam(learning_rate=1e-3)
9autoencoder.compile(
10 optimizer=optimizer,
11 loss='mean_squared_error'
12)
13
14H = autoencoder.fit(
15 train_data,
16 train_data,
17 validation_data=(test_data, test_data),
18 batch_size=32,
19 epochs=epochs,
20 verbose=1
21)
1Epoch 1/50
293/93 [==============================] - 3s 14ms/step - loss: 25.9159 - val_loss: 20.1146
3Epoch 2/50
493/93 [==============================] - 1s 8ms/step - loss: 13.6564 - val_loss: 25.2768
5Epoch 3/50
693/93 [==============================] - 1s 12ms/step - loss: 12.9712 - val_loss: 12.1926
7Epoch 4/50
893/93 [==============================] - 1s 11ms/step - loss: 9.5174 - val_loss: 12.3409
9Epoch 5/50
1093/93 [==============================] - 1s 11ms/step - loss: 8.2311 - val_loss: 7.3879
11Epoch 6/50
1293/93 [==============================] - 1s 13ms/step - loss: 6.7613 - val_loss: 6.8327
13Epoch 7/50
1493/93 [==============================] - 1s 9ms/step - loss: 9.9258 - val_loss: 19.2627
15Epoch 8/50
1693/93 [==============================] - 1s 13ms/step - loss: 10.4538 - val_loss: 7.3759
17Epoch 9/50
1893/93 [==============================] - 1s 12ms/step - loss: 7.9410 - val_loss: 15.4545
19Epoch 10/50
2093/93 [==============================] - 1s 12ms/step - loss: 9.6013 - val_loss: 14.4862
21Epoch 11/50
2293/93 [==============================] - 1s 11ms/step - loss: 7.4179 - val_loss: 9.0731
23Epoch 12/50
2493/93 [==============================] - 1s 7ms/step - loss: 9.4254 - val_loss: 12.0747
25Epoch 13/50
2693/93 [==============================] - 1s 6ms/step - loss: 7.7343 - val_loss: 12.8262
27Epoch 14/50
2893/93 [==============================] - 1s 5ms/step - loss: 6.6644 - val_loss: 12.9122
29Epoch 15/50
3093/93 [==============================] - 0s 4ms/step - loss: 6.9548 - val_loss: 6.6565
31Epoch 16/50
3293/93 [==============================] - 1s 12ms/step - loss: 5.5223 - val_loss: 5.3628
33Epoch 17/50
3493/93 [==============================] - 1s 7ms/step - loss: 4.9501 - val_loss: 5.3698
35Epoch 18/50
3693/93 [==============================] - 1s 9ms/step - loss: 4.4375 - val_loss: 5.7800
37Epoch 19/50
3893/93 [==============================] - 0s 5ms/step - loss: 4.5904 - val_loss: 6.7513
39Epoch 20/50
4093/93 [==============================] - 0s 4ms/step - loss: 4.3928 - val_loss: 5.1352
41Epoch 21/50
4293/93 [==============================] - 0s 5ms/step - loss: 4.2025 - val_loss: 5.0055
43Epoch 22/50
4493/93 [==============================] - 1s 6ms/step - loss: 4.6420 - val_loss: 7.2107
45Epoch 23/50
4693/93 [==============================] - 1s 6ms/step - loss: 4.3076 - val_loss: 5.3522
47Epoch 24/50
4893/93 [==============================] - 1s 6ms/step - loss: 5.0197 - val_loss: 5.9515
49Epoch 25/50
5093/93 [==============================] - 0s 5ms/step - loss: 4.8114 - val_loss: 4.7159
51Epoch 26/50
5293/93 [==============================] - 0s 4ms/step - loss: 4.2462 - val_loss: 4.5241
53Epoch 27/50
5493/93 [==============================] - 1s 13ms/step - loss: 3.9700 - val_loss: 7.7623
55Epoch 28/50
5693/93 [==============================] - 1s 5ms/step - loss: 4.1553 - val_loss: 6.0077
57Epoch 29/50
5893/93 [==============================] - 1s 12ms/step - loss: 4.7138 - val_loss: 6.1912
59Epoch 30/50
6093/93 [==============================] - 0s 5ms/step - loss: 4.0352 - val_loss: 5.7618
61Epoch 31/50
6293/93 [==============================] - 1s 6ms/step - loss: 4.2653 - val_loss: 6.5847
63Epoch 32/50
6493/93 [==============================] - 1s 6ms/step - loss: 4.7350 - val_loss: 5.4481
65Epoch 33/50
6693/93 [==============================] - 1s 6ms/step - loss: 4.2043 - val_loss: 4.3257
67Epoch 34/50
6893/93 [==============================] - 1s 8ms/step - loss: 3.7850 - val_loss: 5.3531
69Epoch 35/50
7093/93 [==============================] - 0s 4ms/step - loss: 3.9837 - val_loss: 5.0959
71Epoch 36/50
7293/93 [==============================] - 1s 8ms/step - loss: 4.1637 - val_loss: 5.3485
73Epoch 37/50
7493/93 [==============================] - 1s 6ms/step - loss: 3.9734 - val_loss: 7.6862
75Epoch 38/50
7693/93 [==============================] - 1s 11ms/step - loss: 6.5799 - val_loss: 8.9945
77Epoch 39/50
7893/93 [==============================] - 1s 9ms/step - loss: 5.3310 - val_loss: 5.6758
79Epoch 40/50
8093/93 [==============================] - 1s 9ms/step - loss: 4.4076 - val_loss: 6.1393
81Epoch 41/50
8293/93 [==============================] - 1s 11ms/step - loss: 3.8459 - val_loss: 4.7130
83Epoch 42/50
8493/93 [==============================] - 1s 9ms/step - loss: 3.7842 - val_loss: 4.5664
85Epoch 43/50
8693/93 [==============================] - 1s 6ms/step - loss: 3.6916 - val_loss: 5.1655
87Epoch 44/50
8893/93 [==============================] - 1s 7ms/step - loss: 3.5001 - val_loss: 3.7929
89Epoch 45/50
9093/93 [==============================] - 1s 11ms/step - loss: 3.3153 - val_loss: 5.3740
91Epoch 46/50
9293/93 [==============================] - 1s 6ms/step - loss: 3.4496 - val_loss: 4.0961
93Epoch 47/50
9493/93 [==============================] - 0s 5ms/step - loss: 3.4558 - val_loss: 4.4008
95Epoch 48/50
9693/93 [==============================] - 1s 11ms/step - loss: 3.4304 - val_loss: 4.9327
97Epoch 49/50
9893/93 [==============================] - 1s 11ms/step - loss: 3.4879 - val_loss: 3.9795
99Epoch 50/50
10093/93 [==============================] - 1s 11ms/step - loss: 3.1195 - val_loss: 4.5895
1plt.style.use("ggplot")
2N = len(H.history["loss"])
3plt.plot(np.arange(1, N + 1), H.history["loss"], label="Loss")
4plt.plot(np.arange(1, N + 1), H.history["val_loss"], label="val_loss")
5plt.xlabel("Epoch #")
6plt.ylabel("Loss")
7plt.legend()
8plt.show()
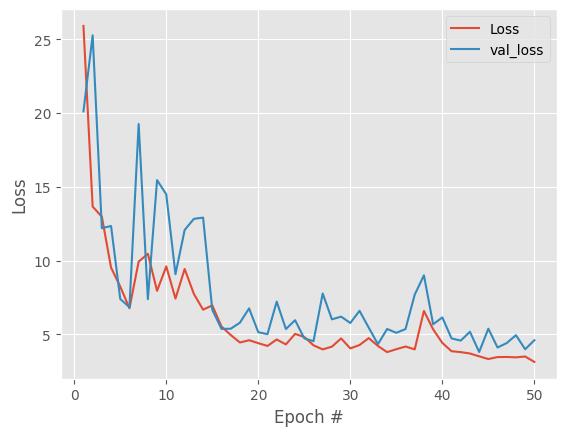
decreasing the learning rate.
1autoencoder = create_l2_noise_autoencoder(
2 neurons,
3 train_data.shape[1],
4 l2=1e-5,
5 noise_std=0.1
6)
7
8optimizer = tf.keras.optimizers.Adam(learning_rate=3e-4)
9autoencoder.compile(
10 optimizer=optimizer,
11 loss='mean_squared_error'
12)
13
14H = autoencoder.fit(
15 train_data,
16 train_data,
17 validation_data=(test_data, test_data),
18 batch_size=32,
19 epochs=epochs,
20 verbose=1
21)
1Epoch 1/50
293/93 [==============================] - 3s 11ms/step - loss: 21.4099 - val_loss: 10.3221
3Epoch 2/50
493/93 [==============================] - 1s 10ms/step - loss: 8.7159 - val_loss: 7.0038
5Epoch 3/50
693/93 [==============================] - 1s 6ms/step - loss: 7.1138 - val_loss: 5.6221
7Epoch 4/50
893/93 [==============================] - 1s 11ms/step - loss: 5.6945 - val_loss: 5.7532
9Epoch 5/50
1093/93 [==============================] - 1s 11ms/step - loss: 4.9997 - val_loss: 4.4984
11Epoch 6/50
1293/93 [==============================] - 1s 13ms/step - loss: 4.5960 - val_loss: 4.6049
13Epoch 7/50
1493/93 [==============================] - 1s 11ms/step - loss: 4.3785 - val_loss: 4.9968
15Epoch 8/50
1693/93 [==============================] - 1s 11ms/step - loss: 3.9819 - val_loss: 4.3294
17Epoch 9/50
1893/93 [==============================] - 1s 12ms/step - loss: 3.8565 - val_loss: 3.8351
19Epoch 10/50
2093/93 [==============================] - 1s 8ms/step - loss: 3.8272 - val_loss: 4.0886
21Epoch 11/50
2293/93 [==============================] - 1s 6ms/step - loss: 3.6578 - val_loss: 3.8850
23Epoch 12/50
2493/93 [==============================] - 1s 13ms/step - loss: 4.0234 - val_loss: 4.2146
25Epoch 13/50
2693/93 [==============================] - 1s 16ms/step - loss: 3.6233 - val_loss: 3.9427
27Epoch 14/50
2893/93 [==============================] - 1s 9ms/step - loss: 3.7437 - val_loss: 4.0461
29Epoch 15/50
3093/93 [==============================] - 1s 13ms/step - loss: 3.3131 - val_loss: 3.2467
31Epoch 16/50
3293/93 [==============================] - 1s 6ms/step - loss: 3.0702 - val_loss: 3.2461
33Epoch 17/50
3493/93 [==============================] - 1s 10ms/step - loss: 3.0544 - val_loss: 3.3153
35Epoch 18/50
3693/93 [==============================] - 1s 14ms/step - loss: 3.4376 - val_loss: 4.1842
37Epoch 19/50
3893/93 [==============================] - 1s 10ms/step - loss: 3.2297 - val_loss: 3.2080
39Epoch 20/50
4093/93 [==============================] - 1s 11ms/step - loss: 2.8850 - val_loss: 3.1777
41Epoch 21/50
4293/93 [==============================] - 1s 12ms/step - loss: 2.8051 - val_loss: 2.9751
43Epoch 22/50
4493/93 [==============================] - 1s 11ms/step - loss: 2.7137 - val_loss: 3.0658
45Epoch 23/50
4693/93 [==============================] - 1s 10ms/step - loss: 2.8133 - val_loss: 3.3787
47Epoch 24/50
4893/93 [==============================] - 1s 10ms/step - loss: 3.5090 - val_loss: 3.1847
49Epoch 25/50
5093/93 [==============================] - 1s 9ms/step - loss: 2.7666 - val_loss: 3.1180
51Epoch 26/50
5293/93 [==============================] - 1s 13ms/step - loss: 2.7511 - val_loss: 3.0354
53Epoch 27/50
5493/93 [==============================] - 1s 12ms/step - loss: 2.9722 - val_loss: 3.0035
55Epoch 28/50
5693/93 [==============================] - 1s 12ms/step - loss: 2.7227 - val_loss: 3.0772
57Epoch 29/50
5893/93 [==============================] - 1s 13ms/step - loss: 2.6147 - val_loss: 2.7778
59Epoch 30/50
6093/93 [==============================] - 1s 12ms/step - loss: 2.5109 - val_loss: 3.0036
61Epoch 31/50
6293/93 [==============================] - 1s 13ms/step - loss: 2.6248 - val_loss: 3.1231
63Epoch 32/50
6493/93 [==============================] - 1s 9ms/step - loss: 2.5655 - val_loss: 3.0635
65Epoch 33/50
6693/93 [==============================] - 1s 12ms/step - loss: 2.5141 - val_loss: 2.8732
67Epoch 34/50
6893/93 [==============================] - 1s 14ms/step - loss: 2.8016 - val_loss: 2.9756
69Epoch 35/50
7093/93 [==============================] - 1s 11ms/step - loss: 2.7626 - val_loss: 3.0096
71Epoch 36/50
7293/93 [==============================] - 1s 11ms/step - loss: 2.8892 - val_loss: 3.1106
73Epoch 37/50
7493/93 [==============================] - 1s 11ms/step - loss: 2.7845 - val_loss: 2.9397
75Epoch 38/50
7693/93 [==============================] - 1s 12ms/step - loss: 2.4996 - val_loss: 3.1788
77Epoch 39/50
7893/93 [==============================] - 1s 13ms/step - loss: 2.4722 - val_loss: 2.7988
79Epoch 40/50
8093/93 [==============================] - 1s 10ms/step - loss: 2.3880 - val_loss: 2.6952
81Epoch 41/50
8293/93 [==============================] - 1s 10ms/step - loss: 2.3410 - val_loss: 2.6565
83Epoch 42/50
8493/93 [==============================] - 1s 10ms/step - loss: 2.3656 - val_loss: 3.5490
85Epoch 43/50
8693/93 [==============================] - 1s 11ms/step - loss: 3.1682 - val_loss: 2.7707
87Epoch 44/50
8893/93 [==============================] - 1s 11ms/step - loss: 2.5004 - val_loss: 2.7223
89Epoch 45/50
9093/93 [==============================] - 1s 12ms/step - loss: 2.4328 - val_loss: 2.6165
91Epoch 46/50
9293/93 [==============================] - 1s 10ms/step - loss: 2.2923 - val_loss: 2.8844
93Epoch 47/50
9493/93 [==============================] - 1s 11ms/step - loss: 2.4613 - val_loss: 2.7295
95Epoch 48/50
9693/93 [==============================] - 1s 9ms/step - loss: 2.3293 - val_loss: 2.6597
97Epoch 49/50
9893/93 [==============================] - 1s 13ms/step - loss: 2.2488 - val_loss: 2.5629
99Epoch 50/50
10093/93 [==============================] - 1s 13ms/step - loss: 2.2132 - val_loss: 2.5368
1plt.style.use("ggplot")
2N = len(H.history["loss"])
3plt.plot(np.arange(1, N + 1), H.history["loss"], label="Loss")
4plt.plot(np.arange(1, N + 1), H.history["val_loss"], label="val_loss")
5plt.xlabel("Epoch #")
6plt.ylabel("Loss")
7plt.legend()
8plt.show()
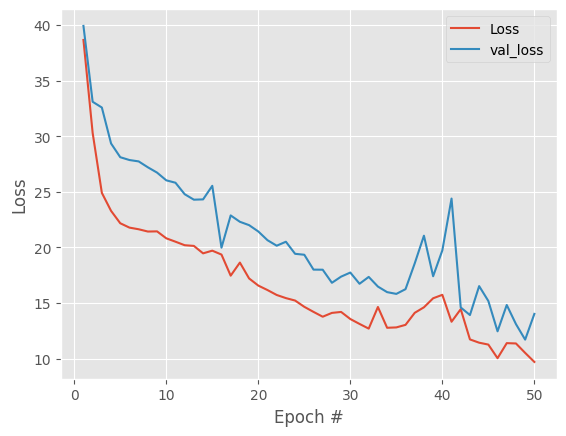
extract and save bottleneck
1hidden_layer_array = np.array(hidden_layer_values)
2hidden_layer_df = pd.DataFrame(hidden_layer_array, columns=[f'Autoencoder_{i}' for i in range(hidden_layer_array.shape[1])])
3csv_output_path = 'hidden_layer.docker.1024.4.hidden50.earlystop.fixPreprocessing.csv'
4hidden_layer_df.to_csv(csv_output_path, index=False)
1csv_output_path = 'hidden_layer.docker.1024.4.hidden50.fixPreprocessing.csv'
2hidden_layer_df.to_csv(csv_output_path, index=False)
add the info to the SCE object
1autoencoder.df <- read.csv( 'hidden_layer.docker.1024.4.hidden50.fixPreprocessing.csv' )
2rownames(autoencoder.df) <- colnames(sce)
3reducedDim( sce, 'AE_50' ) <- autoencoder.df
now plot the results
1plotReducedDim( sce, dimred = 'AE_50', colour_by='clusters' )
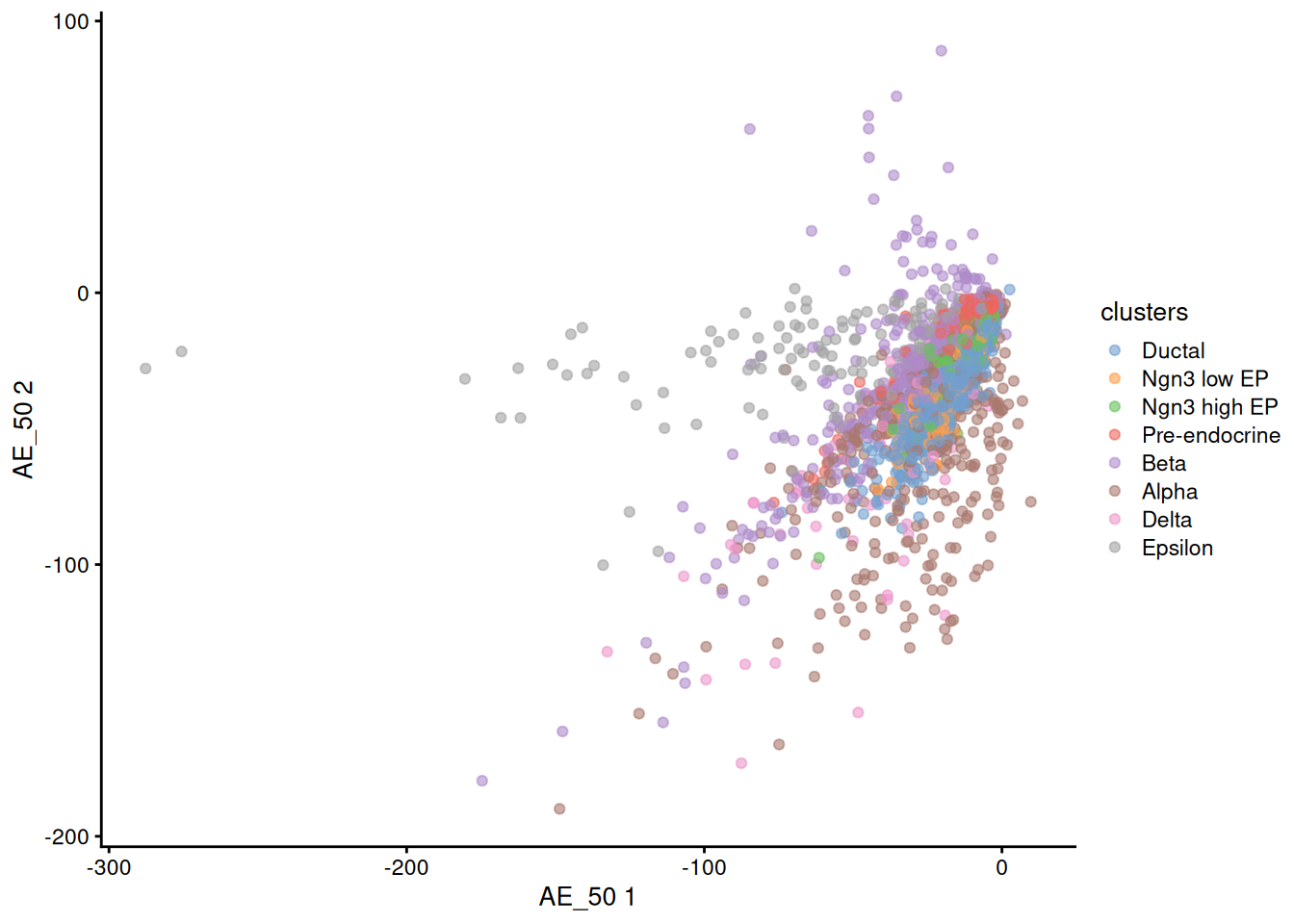
We see something now, it would be interesting to check the rest of dimensions since the neurons of the bottleneck are not arranged by relevance as the PCs.
let's do a tSNE
1sce <- runTSNE( sce, dimred = 'AE_50', name = 'TSNE_AE' )
2plotReducedDim( sce, dimred = 'TSNE_AE', colour_by='clusters' )
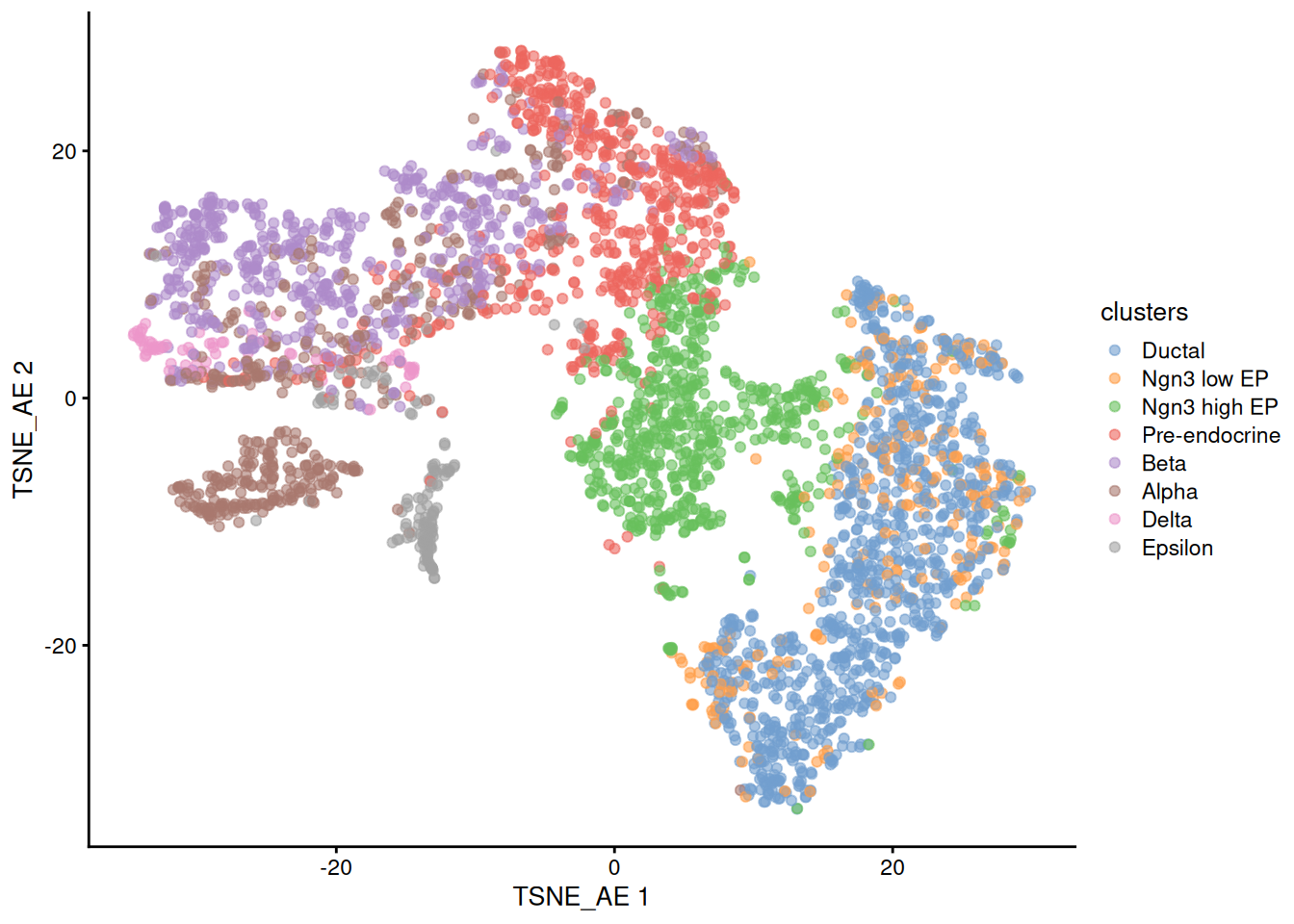
and UMAP
1sce <- runUMAP( sce, dimred = 'AE_50', name = 'UMAP_AE' )
2plotReducedDim( sce, dimred = 'UMAP_AE', colour_by='clusters' )
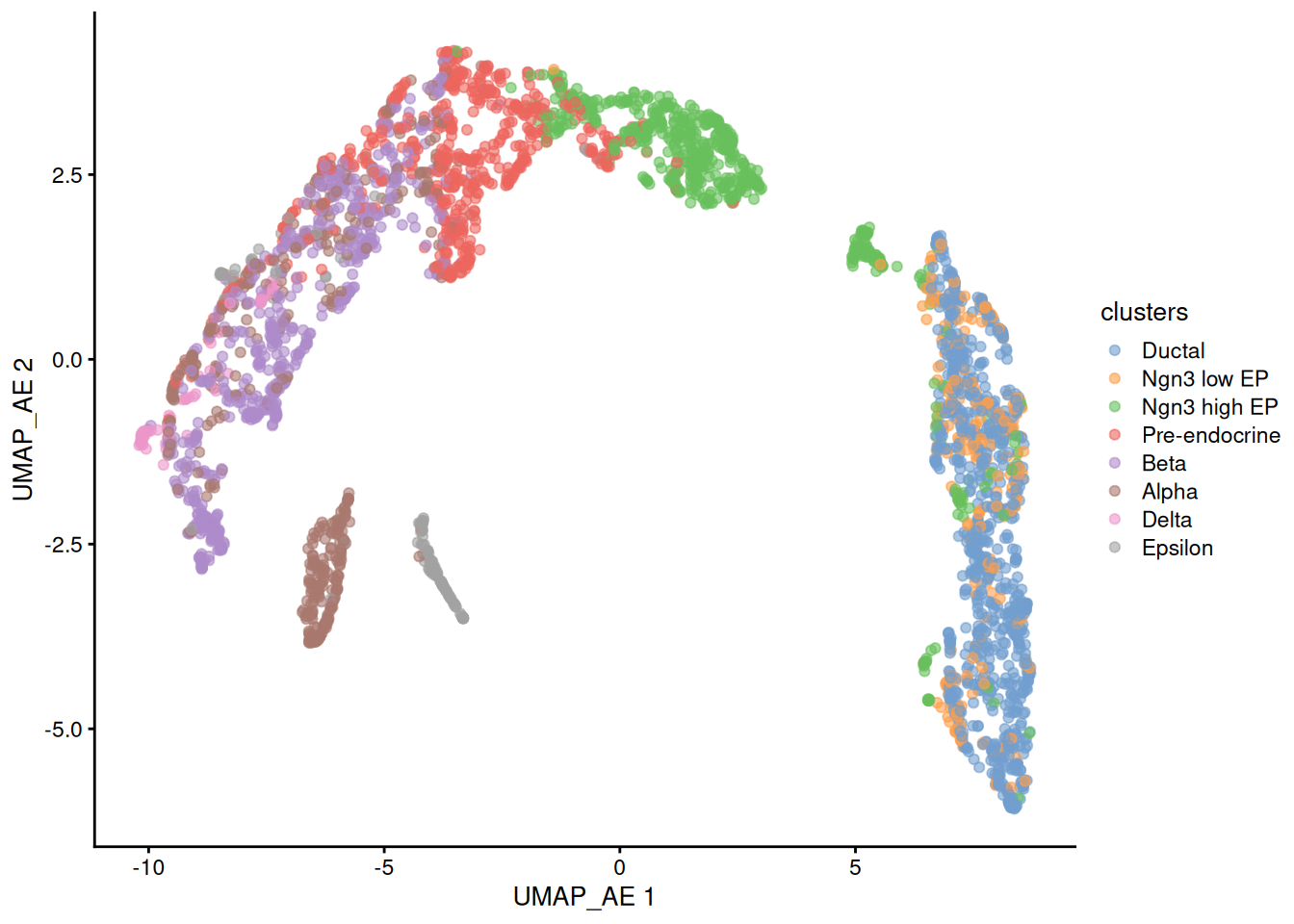
The original UMAP is nicer, but we start to see something in this plot.
Let's try to improve our autoencoder training adding early stopping to prevent overfitting (although it did not seem to be present).
Early stopping is a regularization technique that halts training when the model stops improving, it is useful to prevent overfitting.
Patience defines how many consecutive epochs with no sufficient improvement are allowed before training is stopped.
min_delta specifies the minimum change in the monitored metric that qualifies as an improvement
1from tensorflow.keras.callbacks import EarlyStopping
2early_stop = EarlyStopping(
3 monitor='val_loss',
4 patience=3,
5 min_delta=1e-2,
6 restore_best_weights=True,
7 verbose=1
8)
1autoencoder = create_l2_noise_autoencoder(
2 neurons,
3 train_data.shape[1],
4 l2=1e-5,
5 noise_std=0.1
6)
7
8optimizer = tf.keras.optimizers.Adam(learning_rate=3e-4)
9autoencoder.compile(
10 optimizer=optimizer,
11 loss='mean_squared_error'
12)
1H = autoencoder.fit(
2 x=train_data,
3 y=train_data,
4 validation_data=(test_data, test_data),
5 batch_size=32,
6 epochs=epochs,
7 callbacks=[early_stop],
8 verbose=1
9)
1Epoch 1/50
293/93 [==============================] - 3s 13ms/step - loss: 24.5211 - val_loss: 11.7137
3Epoch 2/50
493/93 [==============================] - 1s 8ms/step - loss: 9.1473 - val_loss: 13.0406
5Epoch 3/50
693/93 [==============================] - 1s 11ms/step - loss: 7.6864 - val_loss: 6.1292
7Epoch 4/50
893/93 [==============================] - 1s 7ms/step - loss: 5.7401 - val_loss: 5.3677
9Epoch 5/50
1093/93 [==============================] - 1s 15ms/step - loss: 5.0120 - val_loss: 4.6072
11Epoch 6/50
1293/93 [==============================] - 1s 10ms/step - loss: 4.4506 - val_loss: 4.4444
13Epoch 7/50
1493/93 [==============================] - 1s 13ms/step - loss: 4.1622 - val_loss: 4.9327
15Epoch 8/50
1693/93 [==============================] - 1s 13ms/step - loss: 4.5532 - val_loss: 4.3296
17Epoch 9/50
1893/93 [==============================] - 1s 12ms/step - loss: 4.1559 - val_loss: 3.8712
19Epoch 10/50
2093/93 [==============================] - 1s 12ms/step - loss: 4.0565 - val_loss: 4.3901
21Epoch 11/50
2293/93 [==============================] - 1s 10ms/step - loss: 3.8009 - val_loss: 3.8640
23Epoch 12/50
2493/93 [==============================] - 1s 8ms/step - loss: 4.2816 - val_loss: 3.7222
25Epoch 13/50
2693/93 [==============================] - 1s 13ms/step - loss: 3.3293 - val_loss: 3.5383
27Epoch 14/50
2893/93 [==============================] - 1s 12ms/step - loss: 3.3962 - val_loss: 3.3154
29Epoch 15/50
3093/93 [==============================] - 1s 11ms/step - loss: 3.1298 - val_loss: 4.0492
31Epoch 16/50
3293/93 [==============================] - 1s 9ms/step - loss: 3.2114 - val_loss: 3.5338
33Epoch 17/50
3492/93 [============================>.] - ETA: 0s - loss: 3.0252Restoring model weights from the end of the best epoch: 14.
3593/93 [==============================] - 1s 11ms/step - loss: 3.0218 - val_loss: 3.5410
36Epoch 17: early stopping
1plt.style.use("ggplot")
2N = len(H.history["loss"])
3plt.plot(np.arange(1, N + 1), H.history["loss"], label="Loss")
4plt.plot(np.arange(1, N + 1), H.history["val_loss"], label="val_loss")
5plt.xlabel("Epoch #")
6plt.ylabel("Loss")
7plt.legend()
8plt.show()
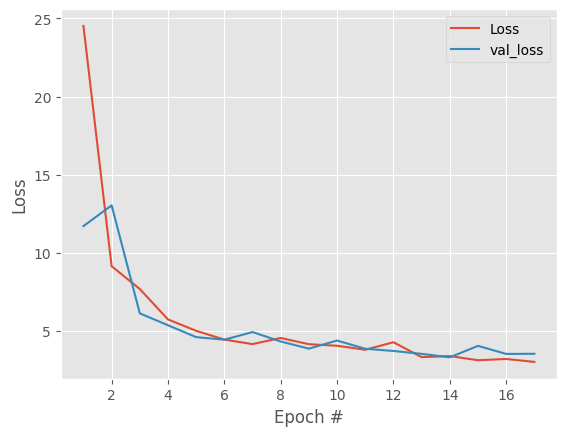
extract and save the bottleneck
1hidden_layer_array = np.array(hidden_layer_values)
2hidden_layer_df = pd.DataFrame(hidden_layer_array, columns=[f'Autoencoder_{i}' for i in range(hidden_layer_array.shape[1])])
3csv_output_path = 'hidden_layer.docker.1024.4.hidden50.earlystop.fixPreprocessing.csv'
4hidden_layer_df.to_csv(csv_output_path, index=False)
add the information to the SCE
1autoencoder.df <- read.csv( 'hidden_layer.docker.1024.4.hidden50.earlystop.fixPreprocessing.csv' )
2rownames(autoencoder.df) <- colnames(sce)
3reducedDim( sce, 'AE_50_early' ) <- autoencoder.df
now plot the results
1plotReducedDim( sce, dimred = 'AE_50_early', colour_by='clusters' )
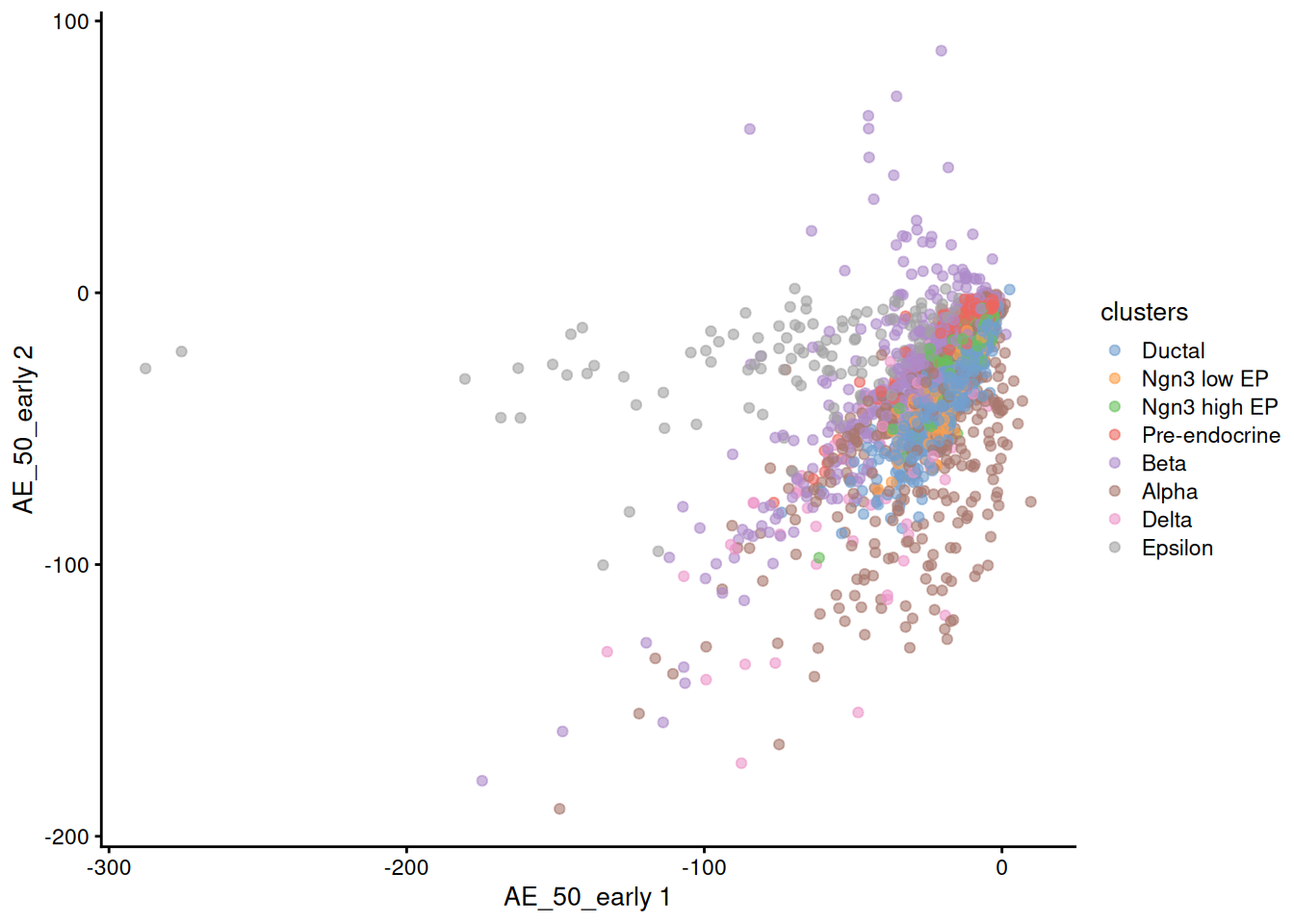
Very similar results, let's see the tSNE
1sce <- runTSNE( sce, dimred = 'AE_50_early', name = 'TSNE_AE2' )
2plotReducedDim( sce, dimred = 'TSNE_AE2', colour_by='clusters' )
and UMAP
1sce <- runUMAP( sce, dimred = 'AE_50_early', name = 'UMAP_AE2' )
2plotReducedDim( sce, dimred = 'UMAP_AE2', colour_by='clusters' )
Again a plot that shows a similar cell organization to the original UMAP although the original representation is still better.
Conclusions
There are several types of autoencoders, today we used the most basic implementation. The most widely used autoencoder type in bioinformatics is the variational autoencoder (VAE), which learns a probability distribution rather than a discrete set of values in the latent space. This probabilistic latent space is well suited to noisy, sparse biological data and supports tasks such as batch correction and data generation. Examples of VAE-based models range from scVI to the more recently described FAVA (Functional Associations using Variational Autoencoders).
Take home messages
- Autoencoders can efficiently compress single cell expression data even with this quick and non-optimal configuaration
- Autoencoders are fast and scalable, making them well suited to the constant increase in dataset size in the single-cell field
- Learning how autoencoders work helps you understand the internal mechanisms of many deep-learning tools, enabling you to better evaluate and interpret their outputs on your own datasets